Code
data <- read_excel("../data/results_survey.xlsx")
data <- data[1:121] %>%
filter(.[[18]] !='Yes') # not analysed any EEG method
font_add_google("Lato")
showtext_opts(dpi = 200)
showtext_auto(enable = TRUE)Here we present researcher’s visualization customs and awareness about some methodological problems.
vec <- c("line", "butterfly", "topo", "topo_map", "topo_array", "erp_image", "parallel", "channel_image")
familiar <- data[61:68] %>% rename_at(vars(colnames(.)), ~ vec) %>%
mutate_at(vars(vec), function(., na.rm = FALSE) (x = ifelse(.=="Yes", 1, 0)))
rec <- data.frame(rowSums(t(familiar))) %>% tibble::rownames_to_column(., "plot") %>%
rename_at(vars(colnames(.)), ~ c("plot", "sum_scores")) %>%
arrange(., desc(sum_scores))
rec %>%
ggplot(data = ., aes(y = reorder(plot, sum_scores), x= sum_scores)) +
geom_col(stat="identity", fill ="lightblue1", col="dodgerblue3") + ylab("plot") + theme_classic() +
geom_text(aes(label = sum_scores, group = plot), position = position_dodge(width = .9), hjust = -0.1) +
theme(legend.position="none", plot.title = element_text(hjust = 0.5)) +
labs(y = "Plot types", x = "Scores") +
ggtitle("Select plots you are familiar with") +
labs(caption = sprintf("N of respondents - %d", nrow(familiar))) +
theme(legend.position="none", plot.caption.position = "plot", plot.caption = element_text(hjust=0), axis.text = element_text(size = 10))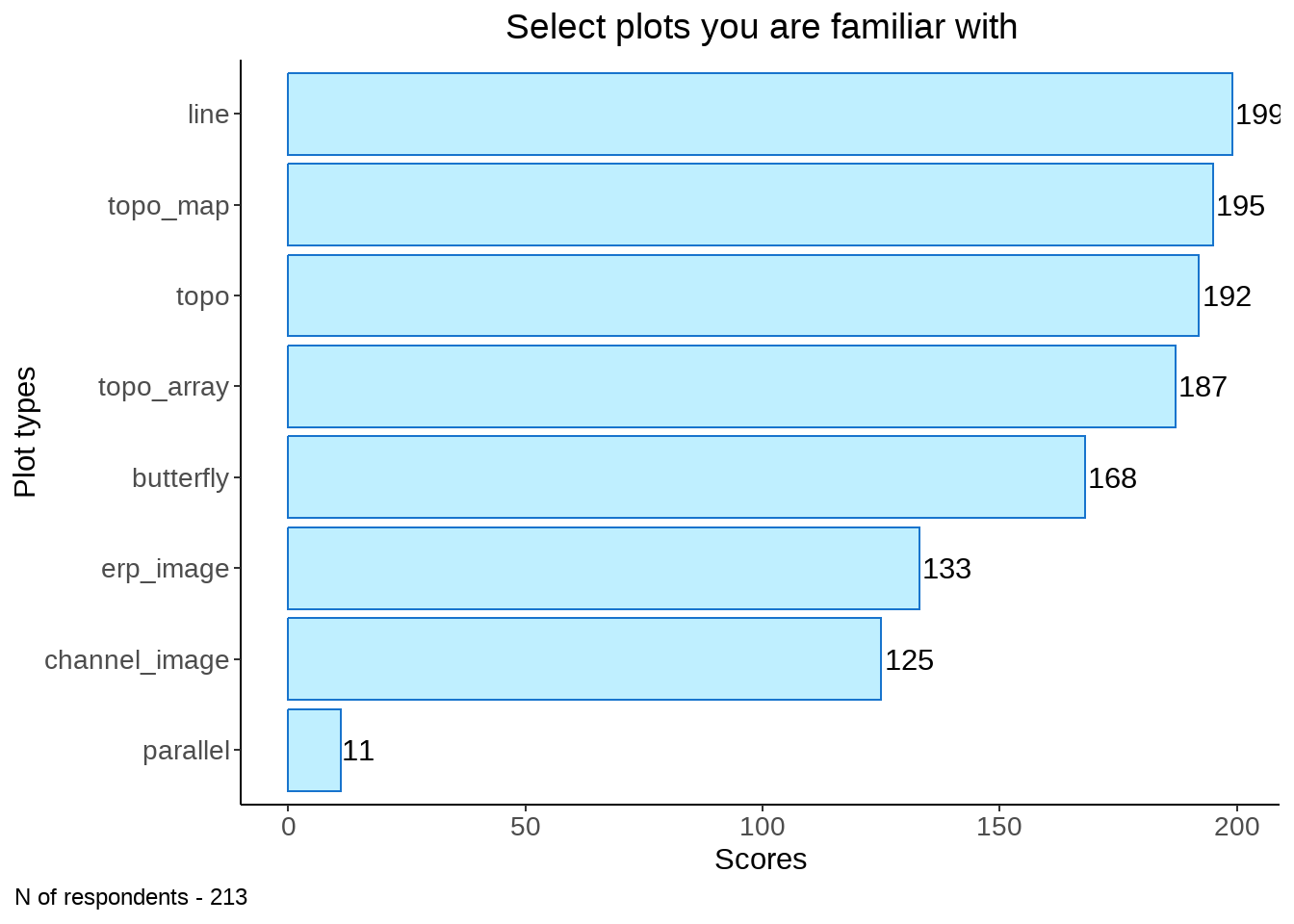
familiar1 <- familiar %>% mutate(n = rowSums(across(where(is.numeric))))
table(familiar1$n) %>% t() %>% data.frame() %>%
mutate(percent_score = round(Freq / 213 * 100)) %>%
ggplot(., aes(y = Var2, x = percent_score)) +
geom_col(stat="identity", fill ="#6BAED6") +
labs(x = "", y = "Number of recognised plots", title = "Number of plots recognised by EEG researchers") + theme_classic() +
geom_text(aes(label = paste0(percent_score, "%")),
position = position_dodge(width = .9), hjust = -0.2, size = 4) + theme(
axis.text.y = element_text(size = 14),
legend.position="none", plot.caption.position = "plot",
plot.caption = element_text(hjust=0),
text = element_text(family = "Lato"),
axis.text.x = element_blank(), axis.text = element_text(size = 10),
plot.title = element_text(color = "grey10", size = 16, face = "bold"),
axis.title.y = element_blank(),
plot.title.position = "plot"
) + xlim(0, 45) 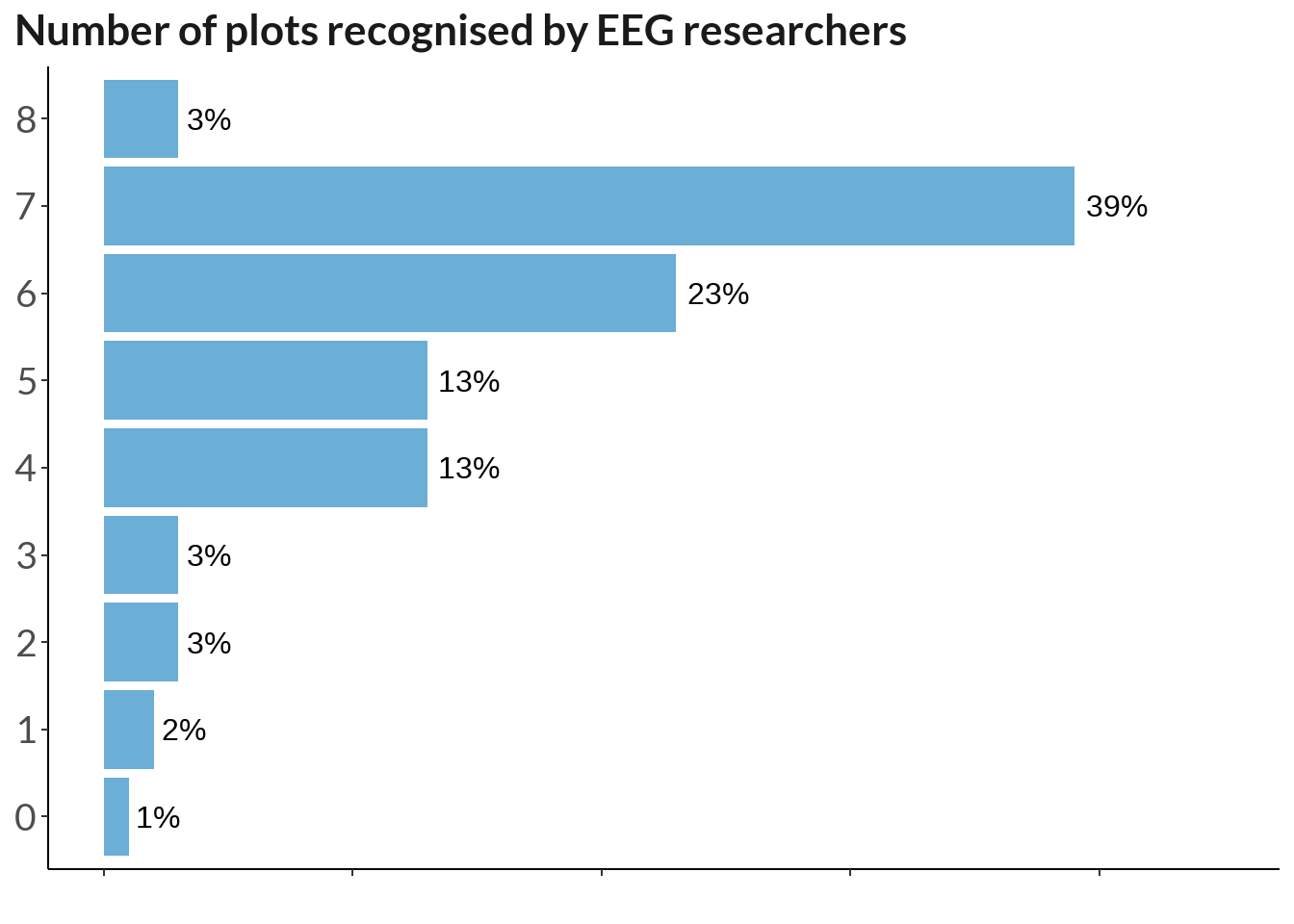
Those who proposed a name for a plot. Bad names are excluded.
stat_preproc <- function(vec){
#N = 70
tmp <- vec %>% filter(!is.na(.)) %>%
dplyr::rename(words = !!names(.)[1]) %>% mutate(words = tolower(words)) %>%
mutate(words = ifelse(nchar(words) < 3, paste(words, "baddd"), words)) %>%
mutate(check =
ifelse(grepl("\\b(baddd|idea|sure|confus|aware|do not|know|why|good|remember|unsure|confusing|mess|unclear|ugly|don't|useless|nan|clear)\\b", words), "bad", "good"))
return(tmp)
}
#stat_preproc(data[vec_named[7]]) %>% View()vec_named <- names(data[ , grepl( "How would you " , names(data))])
plot_names <- c("line", "butterfly", "topo", "topo_series", "erp_grid", "erp_img", "parallel", "channel_img")
na_table <- function(data, vec_named, plot_names){
temp <- data.frame(plot_names)
temp$n <- NA
for (i in 1:8){
n_part <- data[vec_named[i]] %>% stat_preproc(.) %>% #View()
filter(check != "bad") %>%
dplyr::summarise(n = n())
temp$n[i] <- n_part$n
}
return(temp)
}
num_named <- na_table(data, vec_named, plot_names)
num_named %>%
ggplot(., aes(x = n, y = reorder(plot_names, n))) +
geom_col(stat="identity", fill ="#6BAED6") +
labs(x = "Category", y = "Value") +
theme_classic() + theme(axis.title.x=element_blank(), plot.title = element_text(hjust = 0.5)) +
geom_text(aes(label = n), position = position_dodge(width = .9), hjust = -0.1) +
ggtitle("Plot naming") +
labs(caption = sprintf("N of respondents - %d", nrow(familiar))) +
theme(legend.position="none", plot.caption = element_text(hjust=0), axis.text = element_text(size = 10))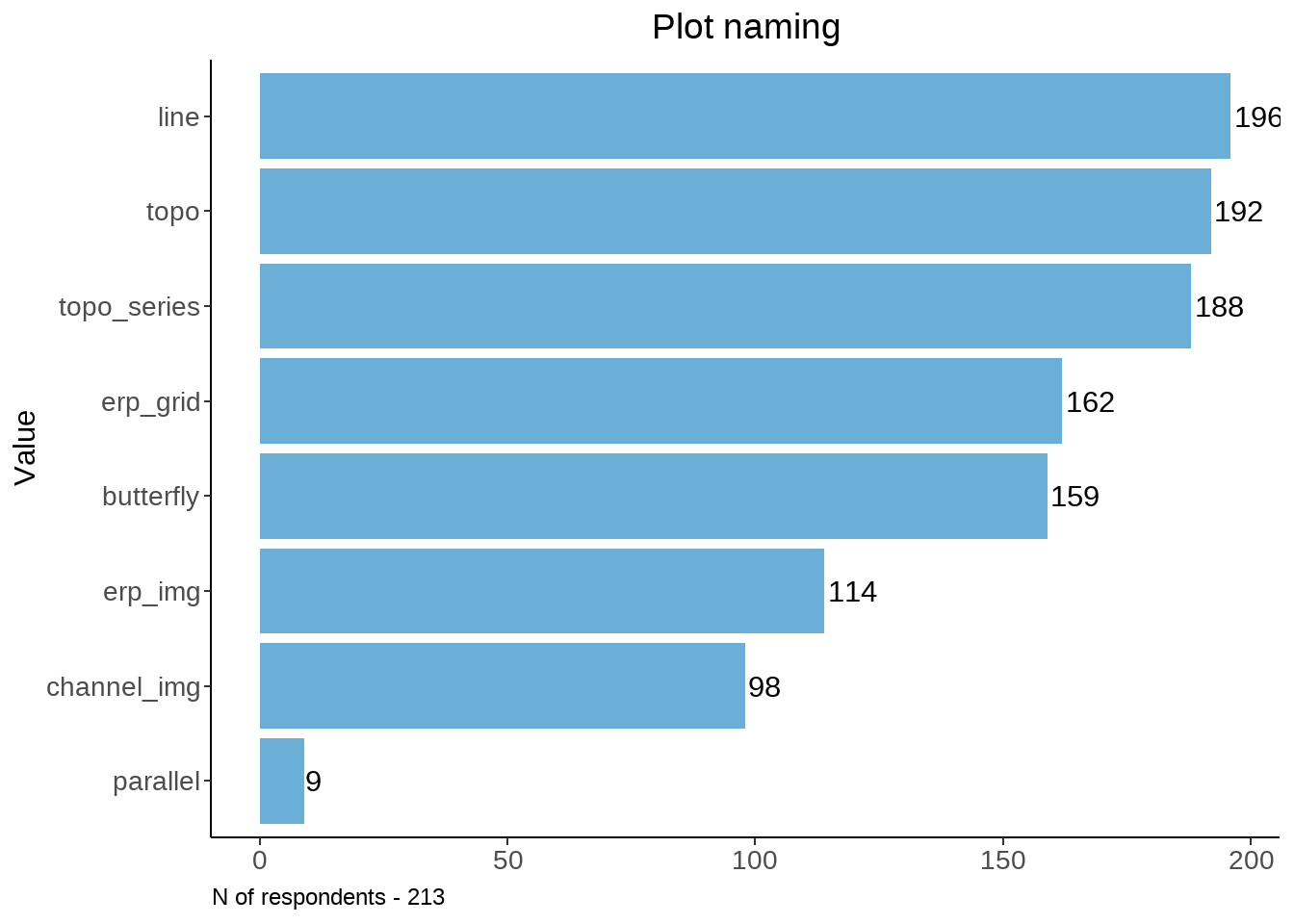
tmp <- data[vec_named]
tmp$n <- rowSums(!is.na(tmp))
table(tmp$n) %>% t() %>% data.frame() %>% select(-Var1) %>%
mutate(percent_score = round(Freq / 213 * 100)) %>%
ggplot(., aes(y = Var2, x = percent_score)) +
geom_col(stat="identity", fill ="#6BAED6") +
labs(x = "", y = "Number of named plots", title = "Number of plots named by EEG researchers") + theme_classic() +
geom_text(aes(label = paste0(percent_score, "%")),
position = position_dodge(width = .9), hjust = -0.2, size = 4) + theme(
axis.text.y = element_text(size = 14),
legend.position="none", plot.caption.position = "plot",
plot.caption = element_text(hjust=0),
text = element_text(family = "Lato"),
axis.text.x = element_blank(), axis.text = element_text(size = 10),
plot.title = element_text(color = "grey10", size = 16, face = "bold"),
axis.title.y = element_blank(),
plot.title.position = "plot"
) + xlim(0, 39) 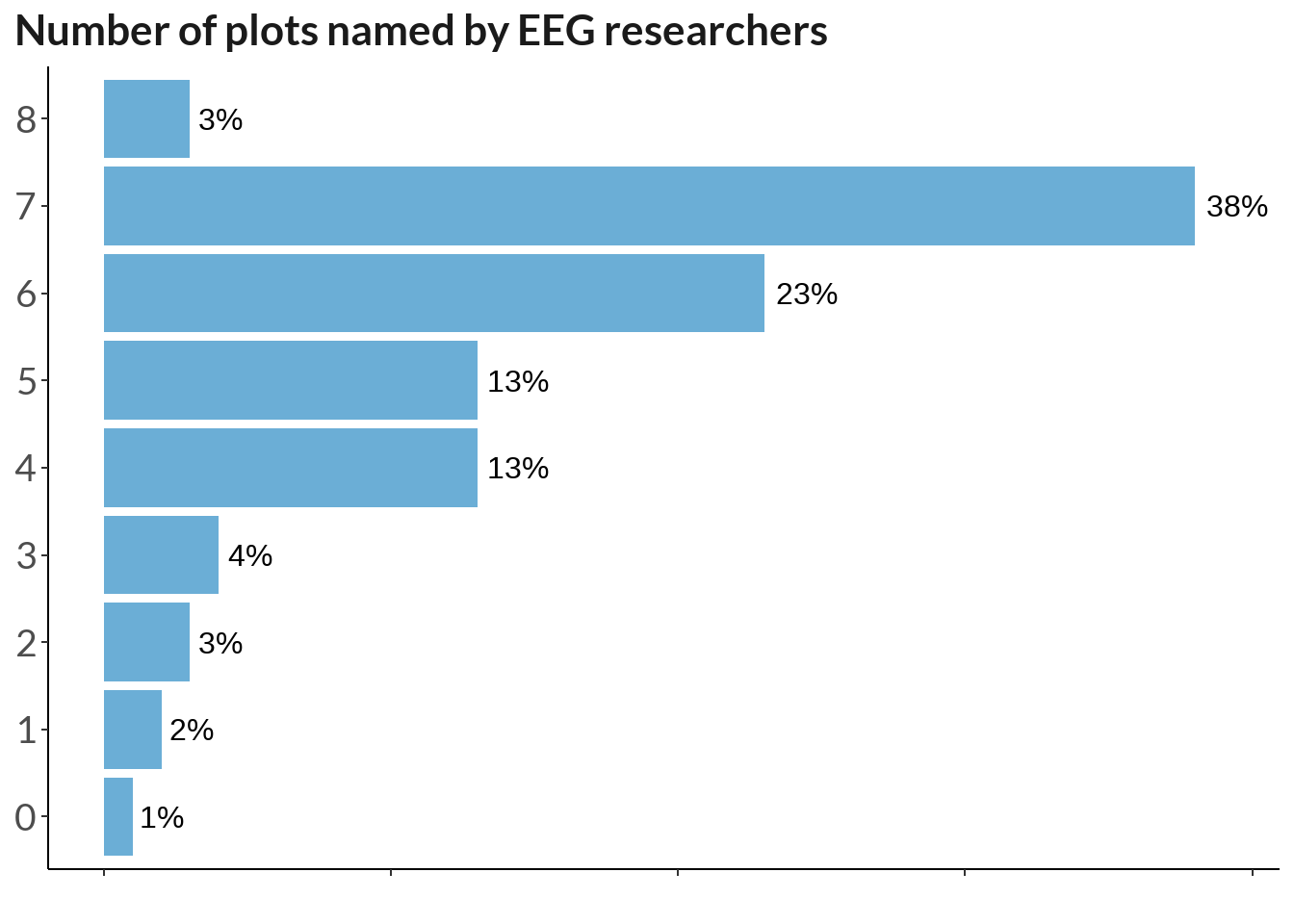
familiar <- data[61:68] %>% rename_at(vars(colnames(.)), ~ plot_names) %>%
mutate_at(vars(plot_names), function(., na.rm = FALSE) (x = ifelse(.=="Yes", 1, 0))) %>%
t() %>% rowSums(.) %>% data.frame(.) %>% tibble::rownames_to_column(., "plot") %>%
rename_at(vars(colnames(.)), ~ c("plots", "recognized"))plot_vec <- rev(c("Parallel\nplot", "Channel\nimage", "ERP image", "Butterfly\nplot", "ERP grid", "Topoplot\ntimeseries", "Topoplot", "ERP plot"))
vec_plotted <- names(data[ , grepl( "Have you ever plotted " , names(data))])
do_vec <- function(vec_plotted, data, plot_names){
t1 <- table(data[vec_plotted[1]])
for (i in 2:length(vec_plotted)) {
t <- table(data[vec_plotted[i]])
t1 <- rbind(t1, t)
}
rownames(t1) <- plot_names
return(t1)
}
tab <- do_vec(vec_plotted, data, plot_names) %>% data.frame() %>% tibble::rownames_to_column(., "plots") %>%
gather(., type, plotted, `N.A`:`Yes`, factor_key=TRUE) %>%
filter(type == "Yes") %>% dplyr::select(-type)
named <- num_named %>% dplyr::rename(named = n, plots = plot_names) mem_tab <- familiar %>%
rename_at(vars(colnames(.)), ~ c("plots", "recognized")) %>%
left_join(., named) %>% left_join(., tab)
mem_tab_fig <- mem_tab %>%
gather(., type, score, recognized:plotted, factor_key=TRUE) %>%
ggplot(., aes(x = score, y = reorder(plots, -score), fill = reorder(type, score))) +
geom_bar(position = "dodge", stat = "identity") +
labs(y = "", x = "Number of users", title = "Level of familiarity with a plot") +
theme_classic() +
geom_text(aes(label = score, group = reorder(type, score)), position = position_dodge(width = .9), hjust = -0.2, size = 3) +
xlim(0, 210) +
scale_fill_brewer(palette = "Blues") +
guides(fill = guide_legend(reverse=T)) +
scale_y_discrete(labels = plot_vec) +
theme(plot.caption = element_text(hjust=0),
axis.text = element_text(size = 10),
text = element_text(family = "Lato"),
legend.title = element_blank(),
legend.position = c(0.8, 0.9),
plot.title = element_text(hjust = 0.5),
axis.title = element_text(size = 12),
plot.title.position = "plot")
mem_tab_fig +
labs(caption = sprintf("N of respondents - %d", nrow(data[61:68])))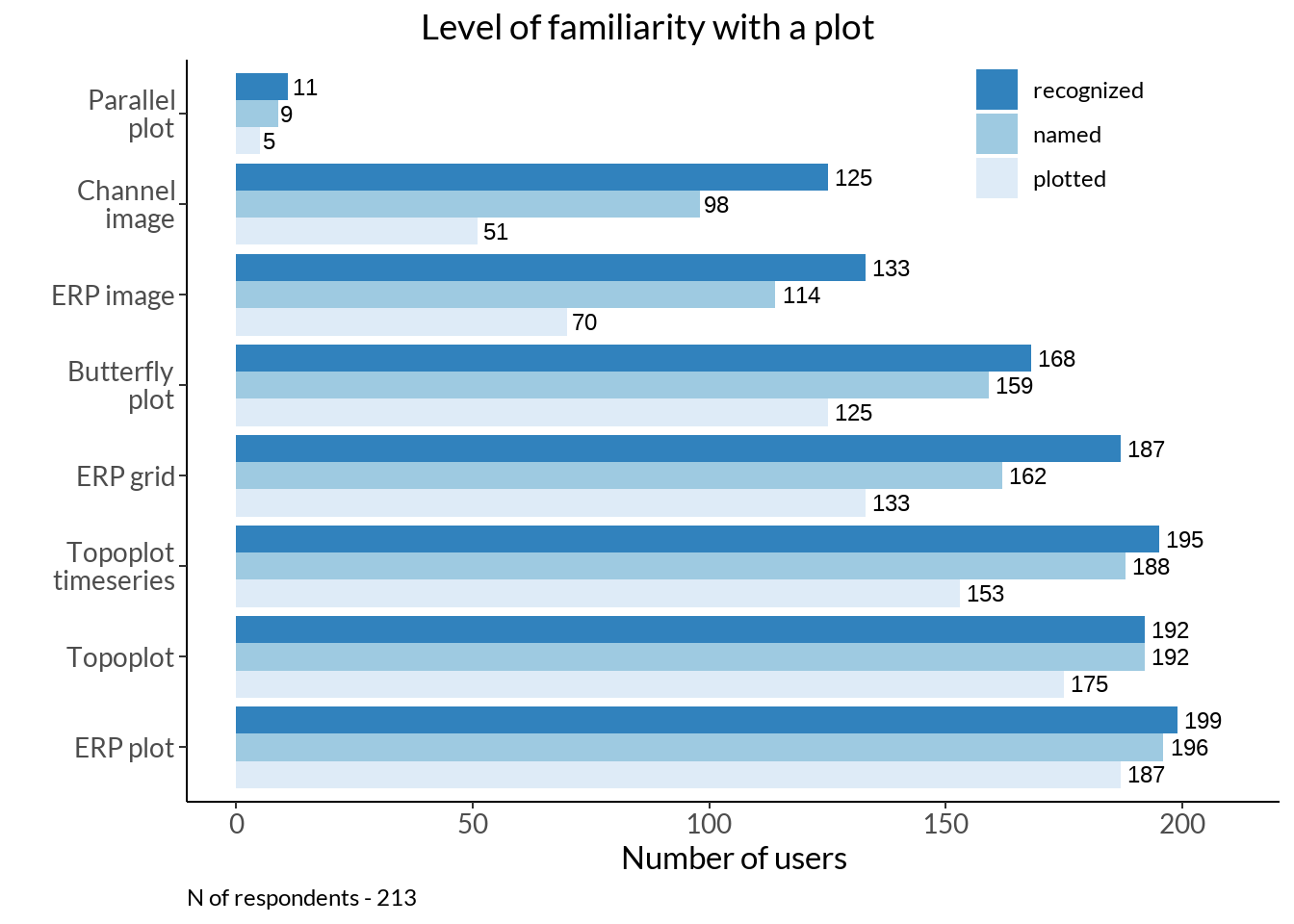
feature <- data[52:60] %>% rename_all(., ~str_split_i(colnames(data[52:60]), "\\? \\[", 2) %>% str_sub(., 1, -2) ) %>%
mutate_at(c(colnames(.)),
funs(recode(.,
"Very important"= 2, "Important"= 1, "Neutral"= 0,
"Low importance"= -1, "Not at all important" = -2 )))
feature %>%
colSums(., na.rm =T) %>% data.frame(.) %>% tibble::rownames_to_column(., "Feature") %>%
filter(!is.na(Feature)) %>%
arrange(desc(.)) %>% rename_at(vars(colnames(.)), ~ c("Feature", "sum_scores")) %>% group_by(Feature) %>%
dplyr::mutate( mean = round(sum_scores / nrow(data), 2)) %>% kbl(escape = F, booktabs = T) %>%
kable_styling("striped", position = "center",) %>% kable_classic(full_width = F, html_font = "Arial")| Feature | sum_scores | mean |
|---|---|---|
| Flexible tweaking of plot attributes (colors, linewidths, margins etc.) | 325 | 1.53 |
| Reproducibility of interactively generated or modified plots | 256 | 1.20 |
| Generating plots by coding | 253 | 1.19 |
| Presentation/publication ready figures | 249 | 1.17 |
| Zooming or panning within a plot | 125 | 0.59 |
| Combine with a custom plot created outside of the toolbox (as subplot or inset) | 121 | 0.57 |
| Speed of plotting | 96 | 0.45 |
| Interactive selection of time-ranges or electrodes e.g. via Sliders or Dropdown menus | 81 | 0.38 |
| Generating plots by clicking (GUI) | -72 | -0.34 |
plot_features <- c(
"Combine with a custom plot created outside of the toolbox (as subplot or inset)",
"Flexible tweaking of plot attributes (colors, linewidths, margins etc.)",
"Speed of plotting",
"Presentation/publication ready figures",
"Reproducibility of interactively generated or modified plots",
"Zooming or panning within a plot",
"Interactive selection of time-ranges or electrodes e.g. via Sliders or Dropdown menus",
"Generating plots by clicking (GUI)",
"Generating plots by coding"
)
comb_data <- feature1 %>% mutate(name = case_when(
name == "Combine with a custom plot created outside of the toolbox (as subplot or inset)" ~ "Inset",
name == "Flexible tweaking of plot attributes (colors, linewidths, margins etc.)"~ "Customizable",
name == "Speed of plotting"~ "Speed",
name == "Presentation/publication ready figures"~ "Publishable",
name == "Reproducibility of interactively generated or modified plots"~ "Reproducible",
name == "Zooming or panning within a plot"~ "Zooming",
name == "Interactive selection of time-ranges or electrodes e.g. via Sliders or Dropdown menus"~ "Interactive",
name == "Generating plots by clicking (GUI)"~ "GUI",
name == "Generating plots by coding" ~ "Coding"
)) %>%
mutate(gr = case_when(
grepl("\\b(Speed|Zooming|GUI|Interactive)\\b", name) == TRUE ~ "Moderate",
grepl("\\b(Coding|Customizable|Reproducible|Publishable|Inset)\\b", name) == TRUE ~ "Favored"))
test <- comb_data %>% group_by(name, value) %>% dplyr::summarise(n = n()) %>%
mutate(gr = case_when(
grepl("\\b(Speed|Zooming|GUI|Interactive|Inset)\\b", name) == TRUE ~ "Moderate",
grepl("\\b(Coding|Customizable|Reproducible|Publishable)\\b", name) == TRUE ~ "Favored")) %>%
mutate(denymax = case_when(value == 2 ~ n, TRUE ~ NA))
cbPalette_fe <- c("#e3342f", "#f6993f", "#f1a20b", "#38c172", "#3490dc", "#6574cd", "#9561e2", "#4dc0b5", "#f66d9b")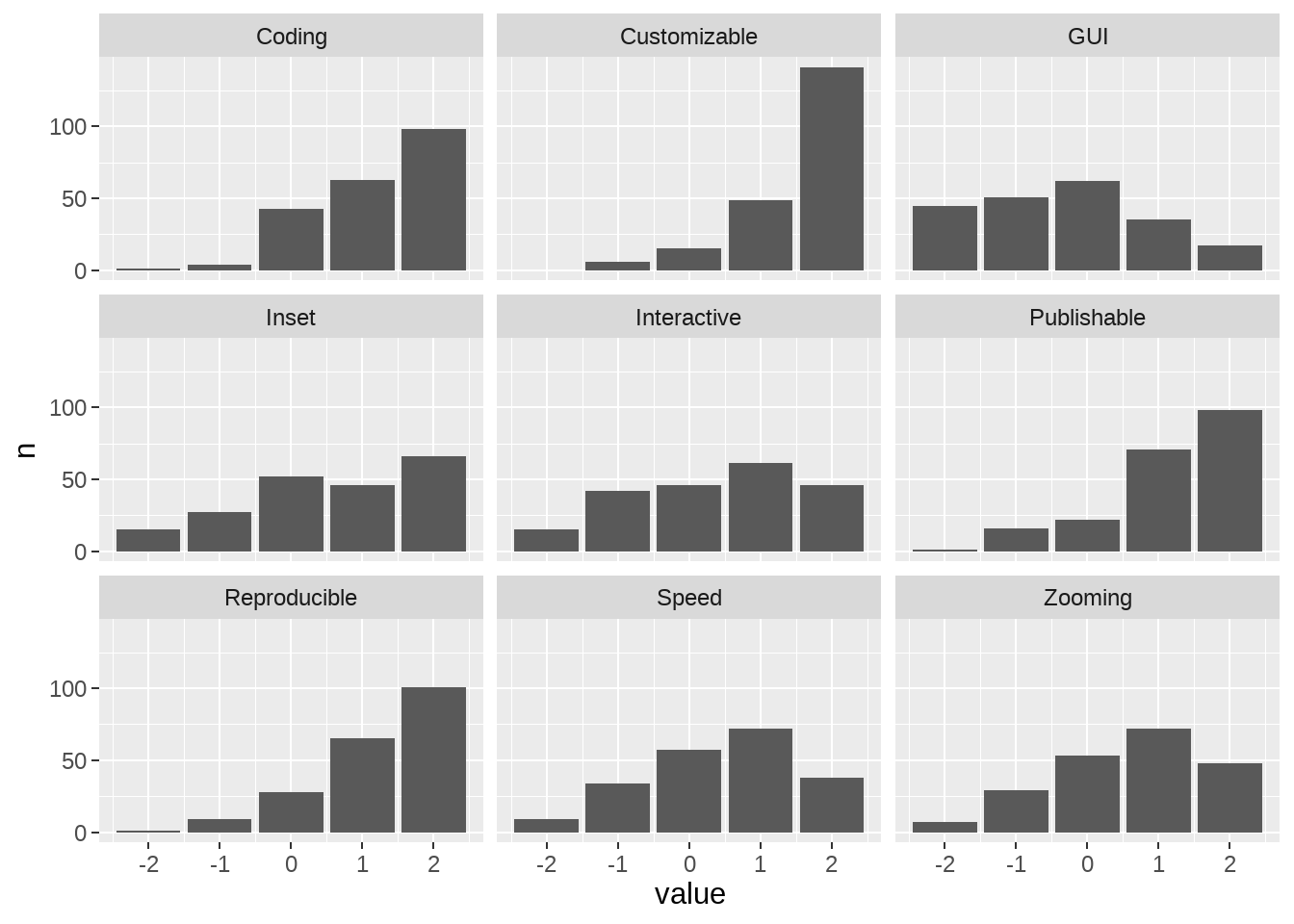
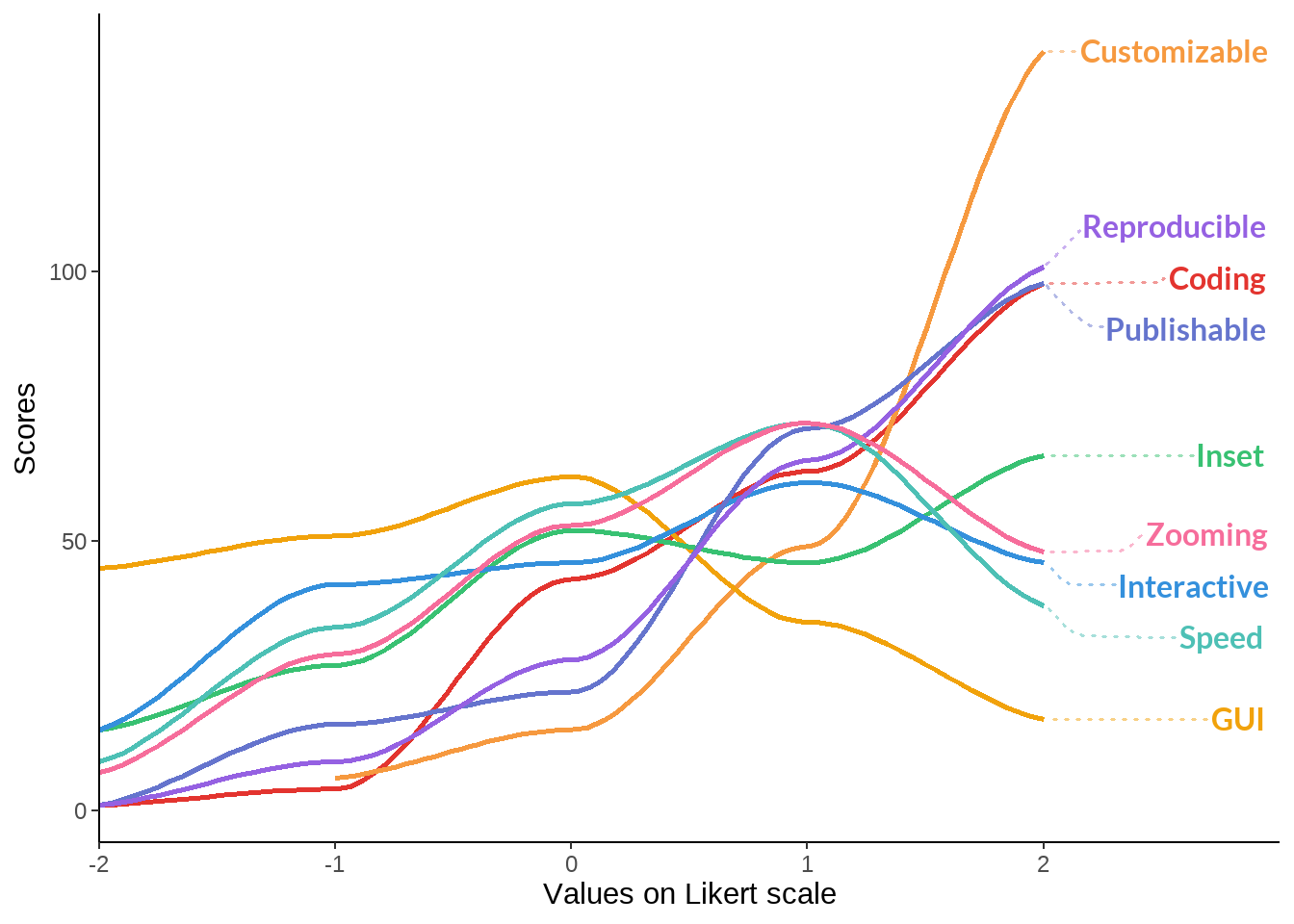
test %>%
ggplot(aes(x = value, y = n, label = name, color = name)) +
geom_smooth(aes(x = value, y = n), size = 1) + #facet_wrap(~gr) +
scale_color_manual(values=cbPalette_fe) + theme_classic() +
theme(legend.position = "none",
strip.background = element_blank(),
strip.text = element_text(size = 14),
) + labs(x = "Values on Likert scale", y = "Scores") +
geom_text_repel(
aes(color = name, label = name, x = 2, y = denymax,),
family = "Lato",
fontface = "bold",
size = 4,
direction = "y",
xlim = c(3, NA),
hjust = 0,
segment.size = .7,
segment.alpha = .5,
segment.linetype = "dotted",
box.padding = .4,
segment.curvature = -0.1,
segment.ncp = 3,
segment.angle = 20
) +
scale_x_continuous(
expand = c(0, 0),
limits = c(-2, 3),
breaks = seq(-2, 2, by = 1)
)
test %>% mutate(value = value + 3) %>%
ggplot(aes(x = value, y = n, label = name, color = name)) +
geom_line(bw = 0.5, size = 1) +
geom_point(shape = 21, fill = 'white', size=2, stroke=1) +
facet_grid(gr~ ., scales = "free_y") +
scale_color_manual(values = cbPalette_fe) +
labs(x = "Values on the Likert scale", y = "Scores") + geom_rangeframe(color = "black") +
theme(
panel.background = element_blank(), panel.border = element_blank(),
legend.position = "none",
text = element_text(family = "Lato"),
strip.background = element_blank(),
#axis.line.x = element_blank(),
axis.text = element_text(color = "grey40"),
axis.ticks = element_line(color = "grey40", size = .5),
strip.text = element_text(size = 14),
axis.title = element_blank(),
plot.title = element_text(
color = "grey10",
size = 16,
face = "bold",
#margin = margin(t = 15)
),
plot.title.position = "plot",
) +
scale_x_continuous(
expand = c(0.01, 0),
limits = c(0.9, 5),
breaks = seq(1, 5, by = 1),
labels = c("Not important", "", "Neutral", "", "Very important")
) +
scale_y_continuous(
expand = c(0.04, 0),
limits = c(0, NA),
breaks = seq(0, 150, by = 20)
) +
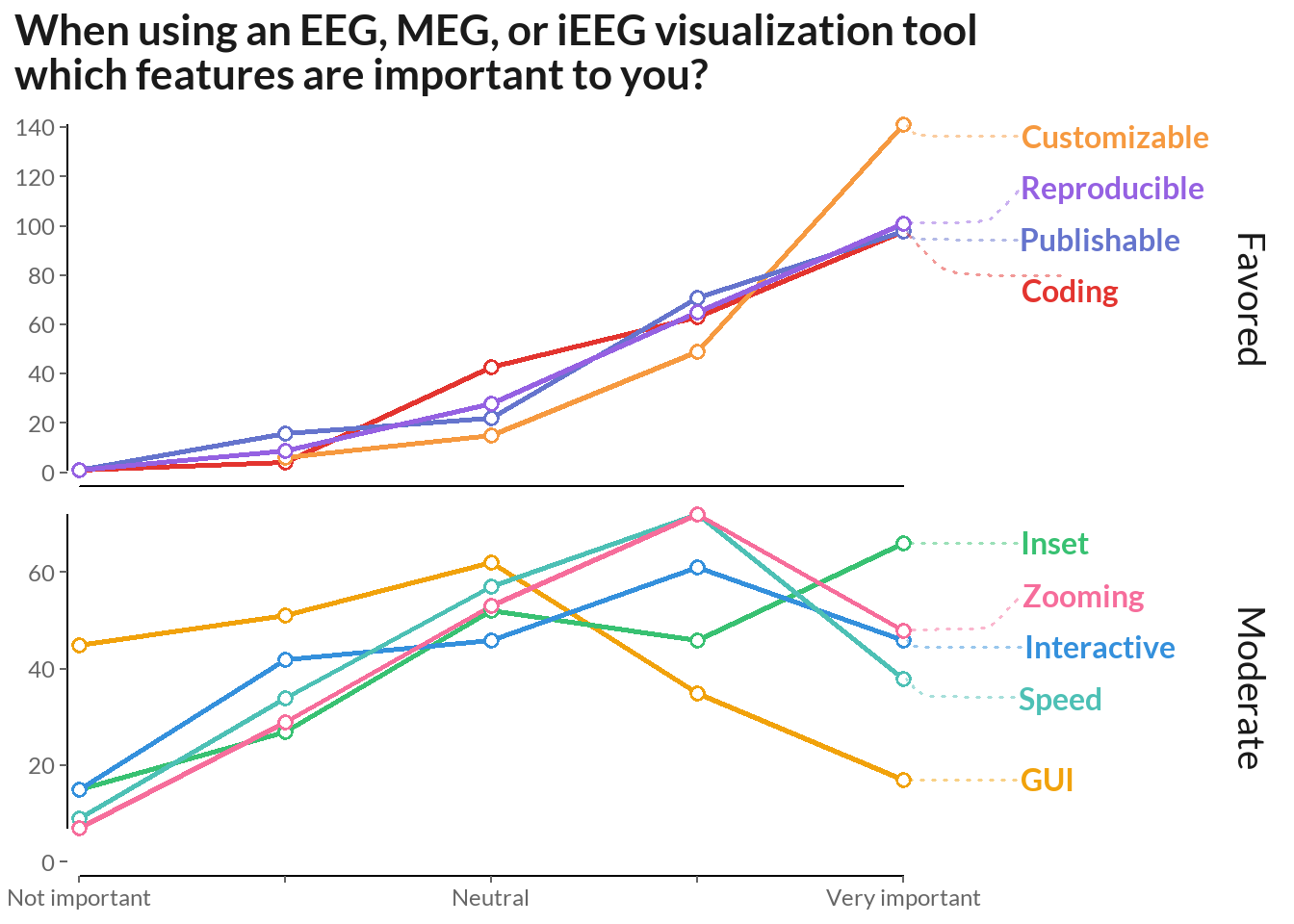
labs(title = "When using an EEG, MEG, or iEEG visualization tool\nwhich features are important to you?") +
coord_cartesian(xlim = c(1, 6.5), clip = "off") +
geom_text_repel(
aes(color = name, label = name, x = 5, y = denymax,),
family = "Lato",
fontface = "bold",
size = 4,
direction = "y",
xlim = c(5.5, NA),
hjust = 0,
segment.size = .7,
segment.alpha = .5,
segment.linetype = "dotted",
box.padding = .4,
segment.curvature = -0.1,
segment.ncp = 3,
segment.angle = 20
)
cbPalette_fe1 <- c("#e3342f", "#6574cd", "#f6993f", "#38c172" )
cbPalette_fe2 <- c("#4dc0b5", "#3490dc","#f1a20b", "#9561e2", "white", "#f66d9b")
core <- function(df){
g <- ggplot(data = df, aes(x = value, y = n, label = name, color = name)) +
geom_line(bw = 0.5, size = 1) +
geom_point(shape = 21, fill = 'white', size=2, stroke=1)+
labs(x = "Values on the Likert scale", y = "Scores") + geom_rangeframe(color = "black") +
theme(
panel.background = element_blank(), panel.border = element_blank(),
legend.position = "none",
text = element_text(family = "Lato"),
strip.background = element_blank(),
axis.text = element_text(color = "grey40"),
axis.ticks = element_line(color = "grey40", size = .5),
strip.text = element_text(size = 14),
axis.title = element_blank(),
plot.title = element_text(
color = "grey10",
size = 16,
face = "bold",
),
plot.title.position = "plot",
) +
coord_cartesian(xlim = c(1, 6.5), clip = "off") +
geom_text_repel(
aes(color = name, label = name, x = 5, y = denymax,),
family = "Lato",
fontface = "bold",
size = 4,
direction = "y",
xlim = c(5.5, NA),
hjust = 0,
segment.size = .7,
segment.alpha = .5,
segment.linetype = "dotted",
box.padding = .4,
segment.curvature = -0.1,
segment.ncp = 3,
segment.angle = 20
)
return(g)
}
test1 <- test %>% filter(gr == "Favored") %>% mutate(value = value + 3) %>%
core(.) +
scale_x_continuous(
expand = c(0.01, 0),
limits = c(0.9, 5),
breaks = seq(1, 5, by = 1)
) +
scale_y_continuous(
expand = c(0.04, 0),
limits = c(0, NA),
breaks = seq(0, 150, by = 30)
) +
scale_color_manual(values = cbPalette_fe1) + theme(axis.text.x=element_blank()) +
labs(title = "When using an EEG, MEG, or iEEG visualization tool\nwhich features are important to you?")
test2 <- test %>% filter(gr != "Favored") %>% mutate(value = value + 3) %>% ungroup() %>%
tibble::add_row(name = "void", value = 1, n = 0, gr = "a", denymax=0) %>%
# line above is just to extend yaxis to zero
core(.) +
scale_x_continuous(
expand = c(0.01, 0),
limits = c(0.9, 5),
breaks = seq(1, 5, by = 1),
labels = c("Not important", "", "Neutral", "", "Very important")
) +
scale_y_continuous(
expand = c(0.04, 0),
limits = c(0, NA),
breaks = seq(0, 72, by = 15)
) +
scale_color_manual(values = cbPalette_fe2) + theme(axis.text.x = element_text(color = "black"))
figure <- ggarrange(test1, test2, align = 'v', nrow = 2)
require(grid)
annotate_figure(figure, left = textGrob("Number of responses", rot = 90, vjust = 1, gp = gpar(cex = 1.3)))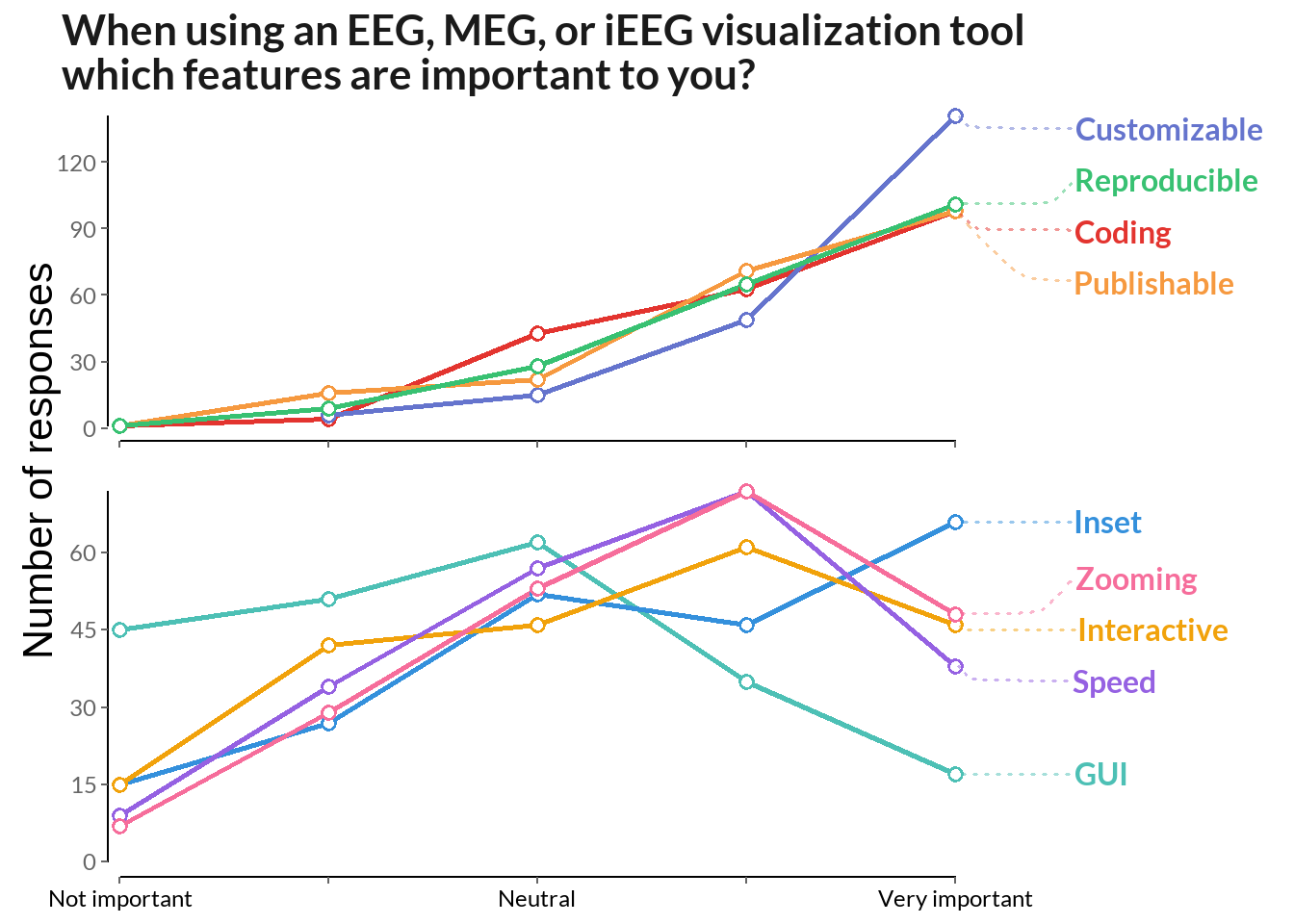
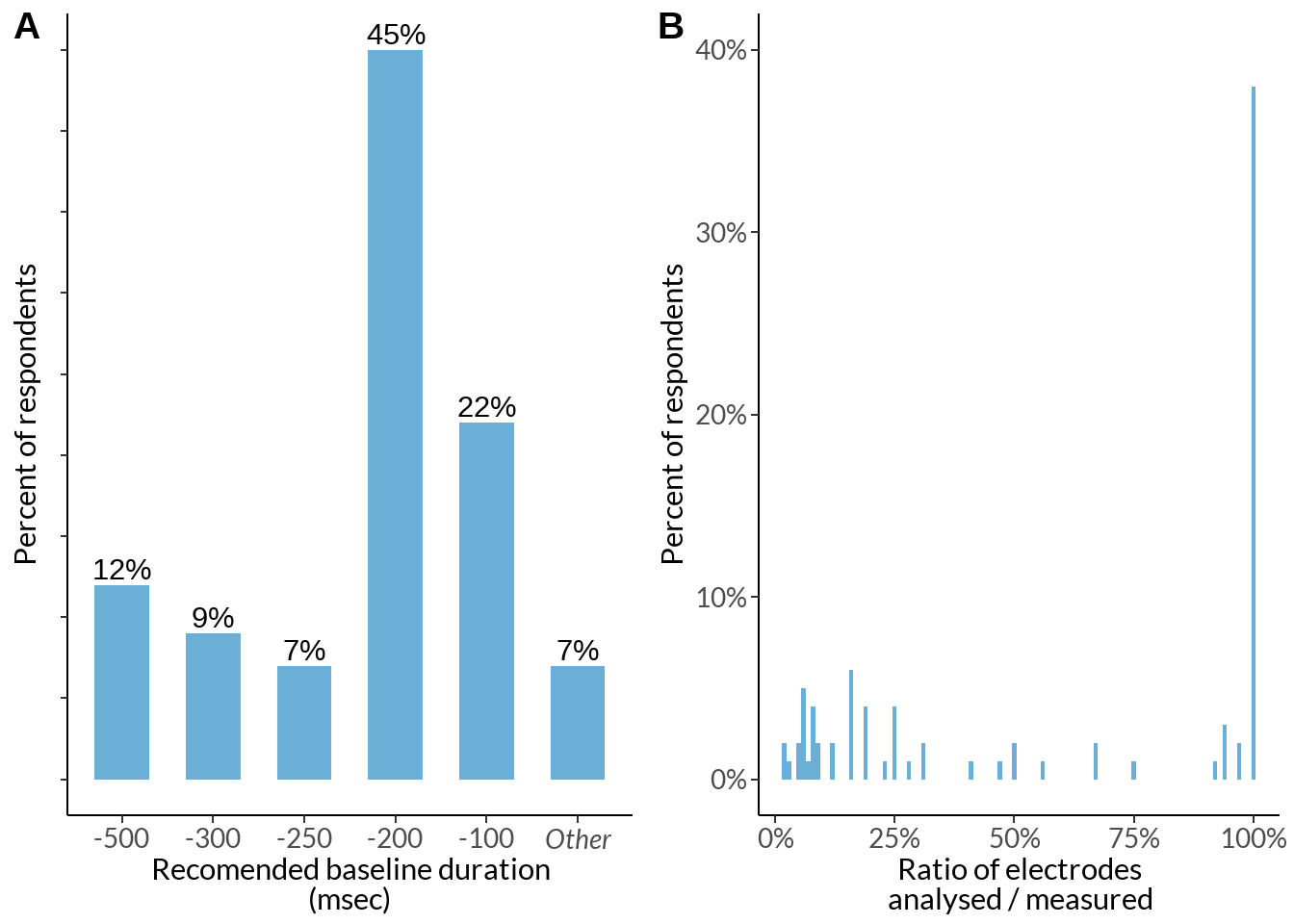
How many channels do you typically analyse and measure?
cv <- data %>% select(23, 24) %>%
rename_at(vars(colnames(.)), ~ c("measure", "analyse")) %>%
filter(measure < 10000, analyse < 500) %>%
mutate(rate = round(analyse / measure, 2)) %>%
group_by(rate) %>% dplyr::summarise(n = n()) %>%
mutate(p = round(n / sum(n), 2))
an_me_plot <- cv %>%
ggplot(aes(x=rate, y = p)) +
geom_col(position = "identity", bins=300, fill ="#6BAED6") +
labs(x ="Ratio of electrodes\nanalysed / measured", y = "") +
theme_classic() +
theme(legend.position="none", plot.caption = element_text(hjust=0), text = element_text(family = "Lato"), axis.text = element_text(size = 10)) +
#ylim(0, 0.1) +
scale_x_continuous(labels = scales::percent) + scale_y_continuous(labels = scales::percent, limits = c(0, 0.4)) +
labs(y = "Percent of respondents")
an_me_plot +
labs(caption = sprintf("N of respondents - %d", sum(cv$n))) 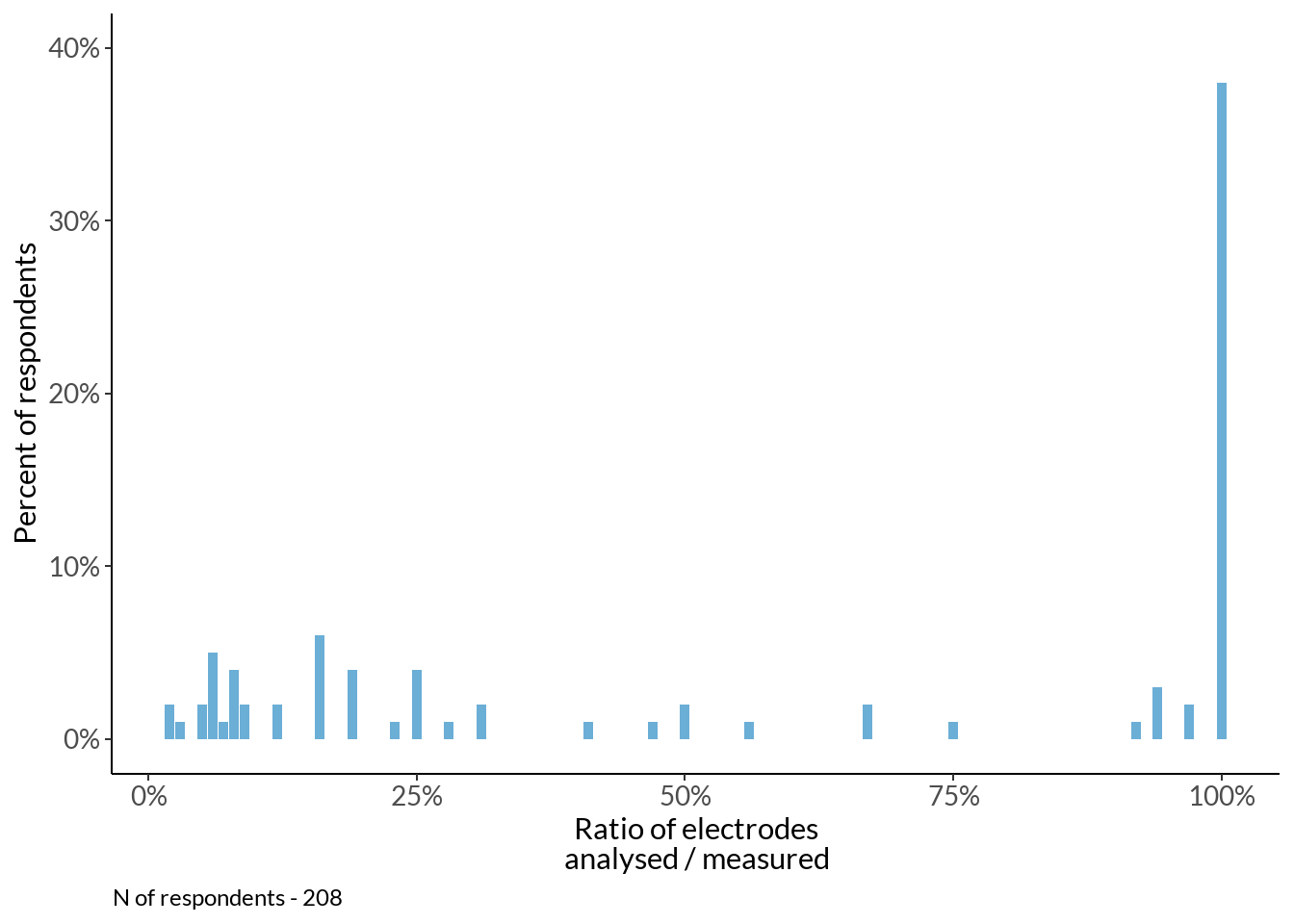
marg <- data %>% select(23, 24) %>%
rename_at(vars(colnames(.)), ~ c("Recorded", "Analysed")) %>%
filter(Recorded < 10000, Analysed < 500) %>%
ggplot(., aes(y=Analysed, x = Recorded)) + geom_point() +
theme(legend.position="none") + theme_classic() + labs(x = "Electrodes recorded", y = "Electrodes aanlysed")
ggMarginal(marg, type="histogram")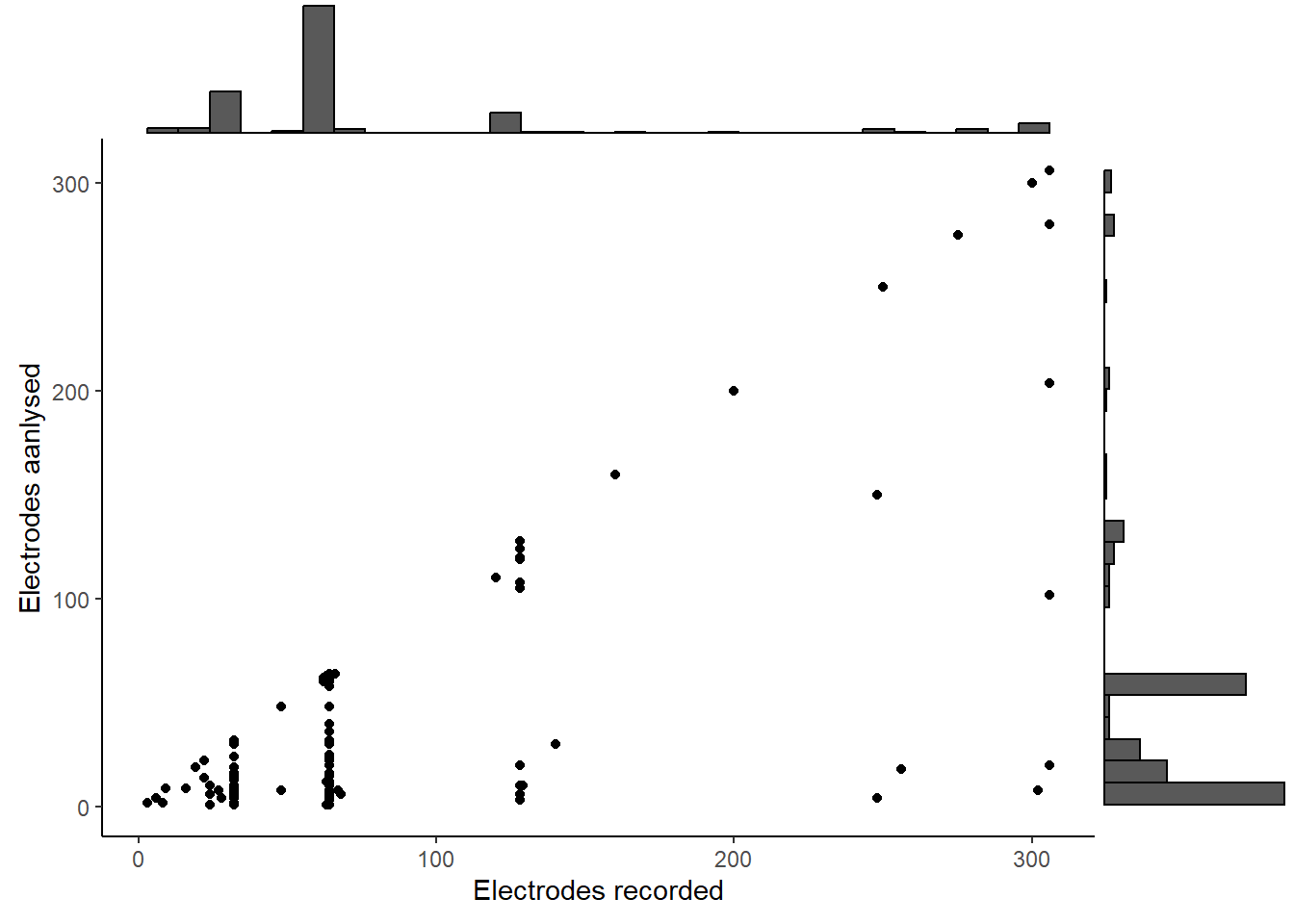
t <- foreach(i = 1:nrow(data)) %do% tokenize_words(as.character(data[i, 11]))
tt <- foreach(i = 1:length(t)) %do% paste(unlist(t[i]), collapse = ' ')
area1 <- data.frame(matrix(tt)) %>% dplyr::rename(words = !!names(.)[1]) %>%
mutate(words = ifelse(str_detect(.[[1]], 'emot|empathy'), "affective neuroscience", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'memory'), "memory", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'hearing|audit'), "auditory", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'cognitive load|selective attention|attention|cognition|consciousness|meditation|cognitive control|self|executive functions'), "attention", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'dbs'), "deep brain stimulation", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'decision making|reward'), "decision making", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'ageing'), "ageing", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'social'), "social cognition", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'olfac'), "olfaction", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'communication|language|speech|biling|english|language processing'), "language and speech", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'bci'), "bci", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'sleep'), "sleep", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'timing|time|temporal'), "time", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'perception'), "perception", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'vis'), "vision", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'developmental|development|ageing'), "development", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'spatial|brain body|motor|motion'), "motor control", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'diagnostics|disorder|psychiatry|epilepsy|autism|patients|therapy|psychopharmacology'), "mental disorders", words)) %>%
mutate(words =ifelse(str_detect(.[[1]], 'signal processing|event related potentials|method|sdf|ieeg'), "dsp", words))
area <- area1 %>%
mutate(words = ifelse(str_detect(.[[1]], 'olfaction|vision|auditory|pain'), "perception", words))%>%
mutate(words = ifelse(str_detect(.[[1]], 'dsp|computational neuroscience'), "methodology", words)) %>%
mutate(words = ifelse(str_detect(.[[1]], 'mental disorders|deep brain stimulation'), "clinical", words))%>%
mutate(words = ifelse(str_detect(.[[1]], 'social cognition'), "affective neuroscience", words))colours = c("#f9a65a", "#599ad3")
ud <- table(data[79]) %>% data.frame() %>%
rename_at(vars(colnames(.)), ~ c("position", "scores")) %>%
mutate(percent_score = round(scores / sum(scores) * 100))
pol_not_plot1 <- ud %>%
ggplot(., aes(y = position, x = percent_score)) + #, fill = as.factor(percent_score))) +
geom_bar(position = "dodge", stat = "identity", width=0.5, fill ="lightblue1", colour ="dodgerblue3") +
theme_classic() +
theme(axis.title=element_blank(),
legend.position="none", axis.text.x = element_blank(),
axis.text = element_text(size = 12),
plot.title = element_text(hjust = 0.5)) +
geom_text(aes(label = paste0(percent_score, "%"), group = position), hjust = -0.2) +
ggtitle("In ERP plot, should positive voltages be plotted upwards, or downwards?") +
scale_fill_manual(values=colours) + xlim(0, 83) +
theme(legend.position="none", plot.caption = element_text(hjust=0), axis.text = element_text(size = 10))
pol_not_plot1 +
labs(caption = sprintf("N of respondents - %d", sum(ud$scores))) 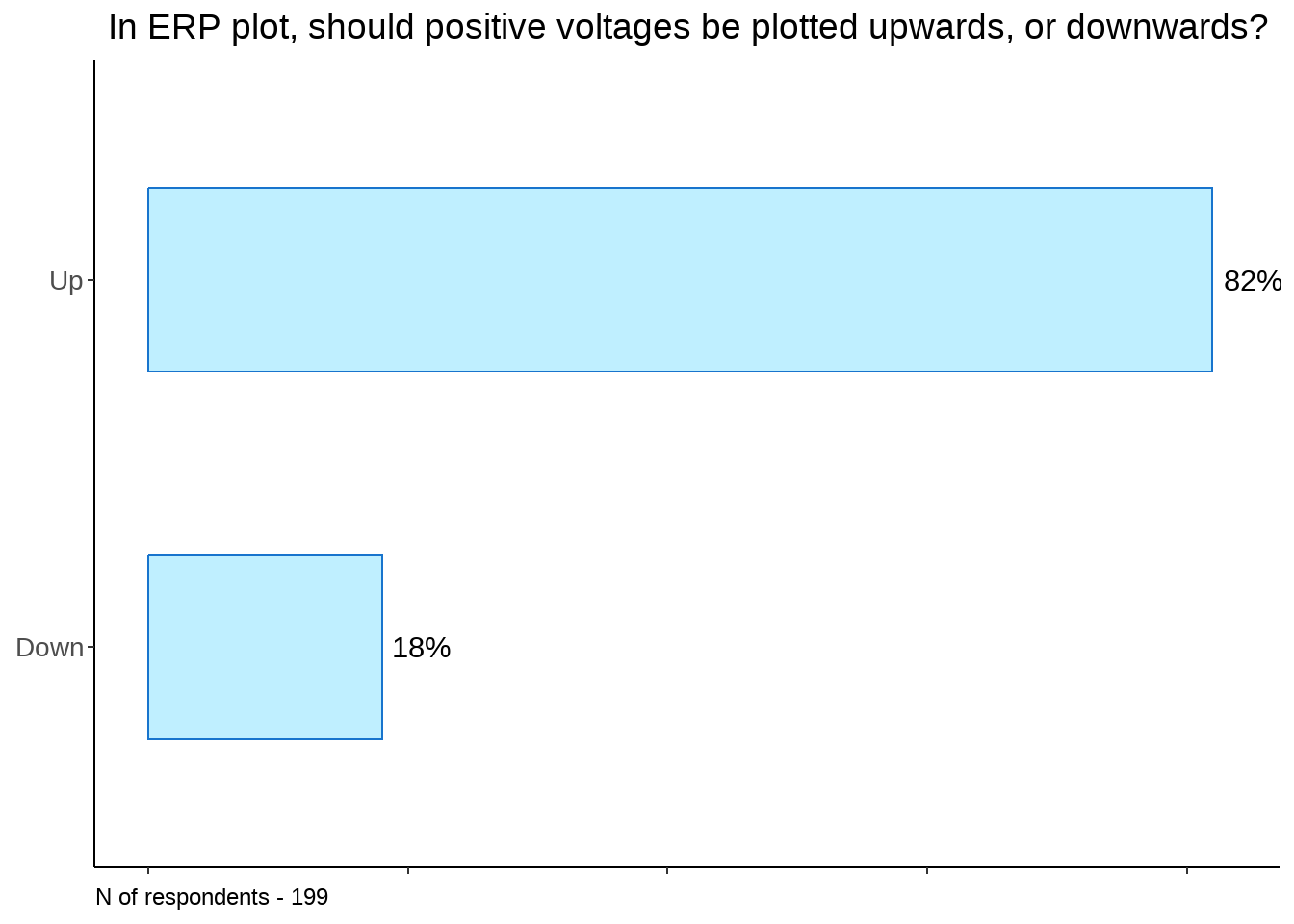
as <- cbind(unlist(area$words), unlist(data[[79]])) %>% data.frame() %>%
filter(.[[2]] != "NANA") %>% dplyr::rename(area=X1, ud=X2) %>%
group_by(area) %>%
filter(n() > 2) %>% group_by(area, ud) %>% dplyr::summarise(n = n()) %>%
group_by(area) %>% dplyr::mutate(nn = sum(n)) %>% filter(area != "")
pol_not_plot2 <- as %>%
ggplot(., aes(x = n, y = reorder(area, nn), fill = ud)) +
geom_col(stat = "identity", width = 0.7) +
labs(x = "Percent of respondents", y = "", fill ="Positive:", title = "In ERP plot, should positive voltages\nbe plotted upwards, or downwards?") +
theme_classic()+
geom_text(aes(label = n, group = ud),
position = position_stack(vjust = 0.5), size = 4) +
coord_cartesian(clip = "off") +
theme(axis.text = element_text(size = 12),
text = element_text(family = "Lato"),
axis.title = element_text(size = 12),
title = element_text(size = 14),
axis.title.y=element_blank(),
legend.title = element_blank(),
legend.direction = "horizontal",
legend.position = c(0.8, 0.2),
plot.title.position = "plot",
plot.caption.position = "plot",
) +
scale_fill_manual(values=colours, limits = c("Up", "Down"))
pol_not_plot2 +
labs(caption = sprintf("N of respondents - %d", sum(as$n)))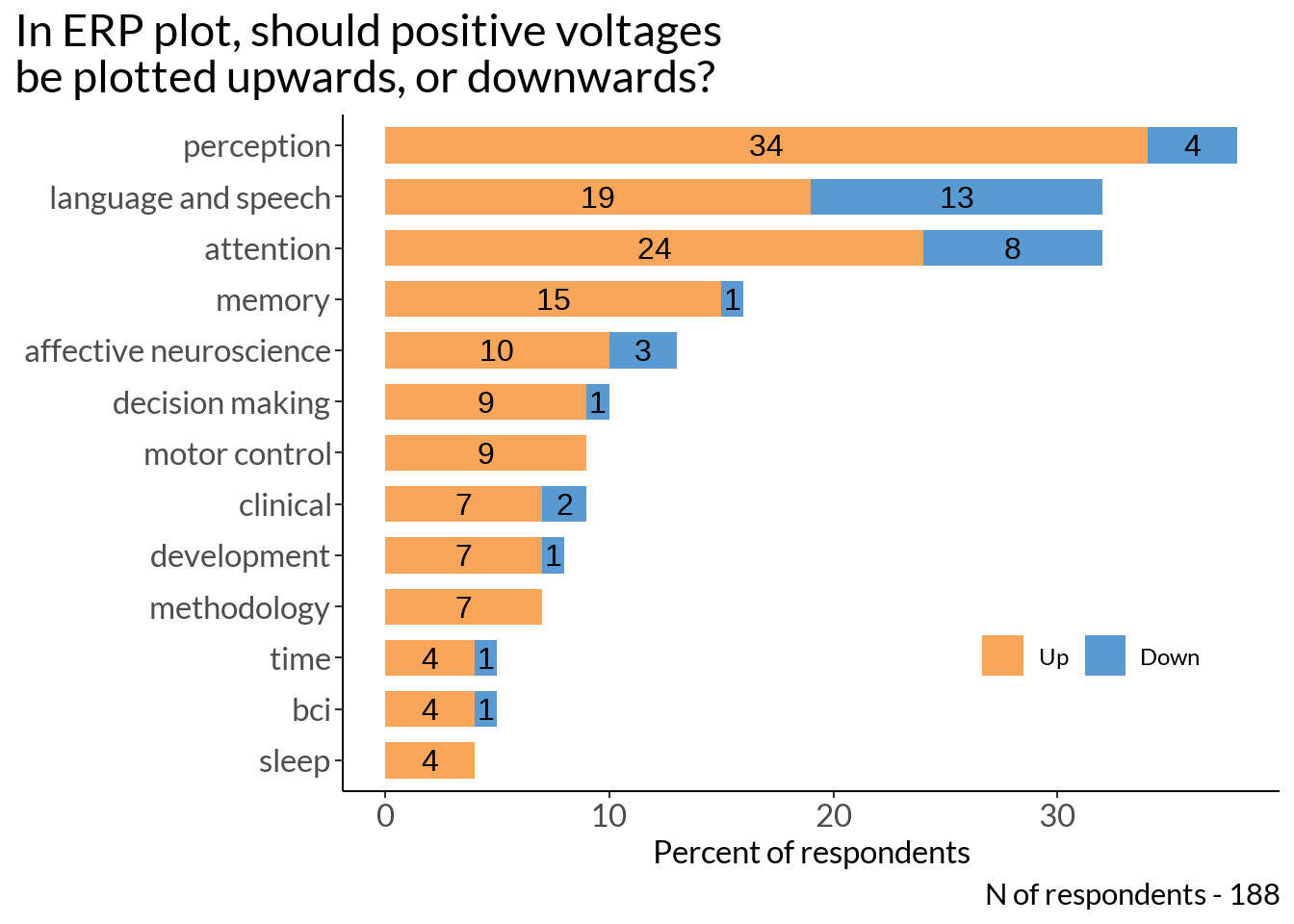
area_short <- area %>%
mutate(words = ifelse(str_detect(.[[1]], 'language and speech|attention'),
"language, speech,
attention",
"affective neuroscience,\nbci, clinical, decision making,\ndevelopment, memory,\nmethodology, motor control,\nperception, sleep, time"))
as2 <- cbind(unlist(area_short$words), unlist(data[[79]])) %>% data.frame() %>%
filter(.[[2]] != "NANA") %>% dplyr::rename(area=X1, ud=X2) %>%
group_by(area) %>%
filter(n() > 2) %>% group_by(area, ud) %>% dplyr::summarise(n = n()) %>%
group_by(area) %>% dplyr::mutate(nn = sum(n)) %>% filter(area != "")
pol_not_plot3 <- as2 %>%
ggplot(., aes(x = n, y = reorder(area, nn), fill = ud)) +
geom_col(stat = "identity", width = 0.7) +
labs(x = "Percent of respondents", y = "", fill ="Positive:", title = "In ERP plot, should positive voltages\nbe plotted upwards, or downwards?") +
theme_classic()+
geom_text(aes(label = n, group = ud),
position = position_stack(vjust = 0.5), size = 4) +
coord_cartesian(clip = "off") +
theme(axis.text = element_text(size = 12),
text = element_text(family = "Lato"),
axis.title = element_text(size = 12),
title = element_text(size = 14),
axis.title.y=element_blank(),
legend.title = element_blank(),
legend.direction = "horizontal",
legend.position = c(0.8, 0.2),
plot.title.position = "plot",
plot.caption.position = "plot",
) +
scale_fill_manual(values=colours, limits = c("Up", "Down"))
pol_not_plot3 +
labs(caption = sprintf("N of respondents - %d", sum(as$n)))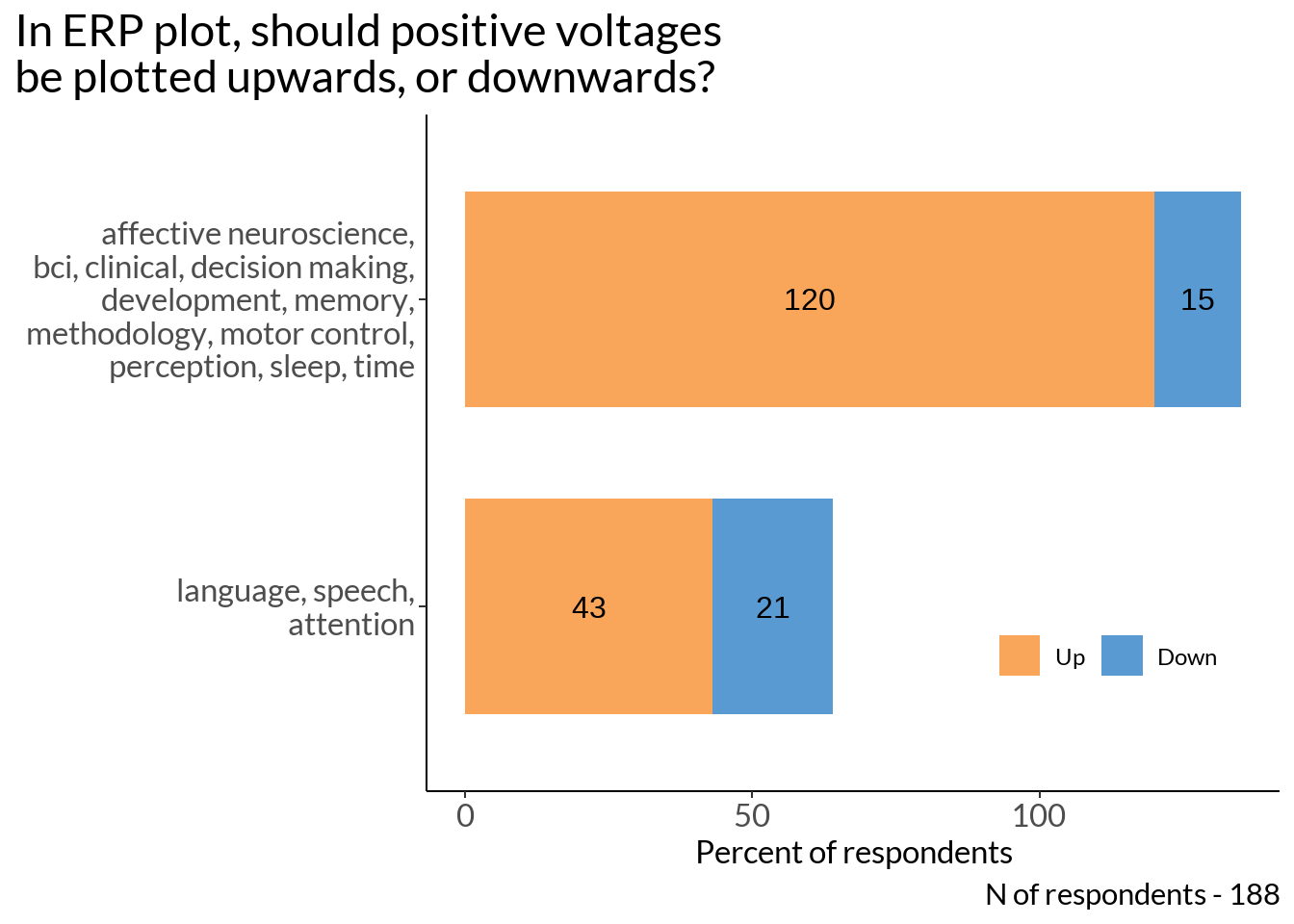
Think about the baseline period (the time before the stimulus onset). How many milliseconds would you recommend to plot? Help: Please, provide the baseline duration for the plot, not the duration for the baseline-correction
# If you don't want to provide a number on previous question, please, provide a justification
j <- data %>%
dplyr::rename(q = !!names(.)[78]) %>% filter(!is.na(q)) %>% dplyr::select(q) %>%
mutate(q = tolower(q)) %>% mutate(q = gsub('depends in|depending on', 'depends on', q),
dependson = ifelse(grepl("depends|depend", q), q, NA)) %>%
separate(dependson, into = c("a","b"), sep = "depends on |depend on ") %>%
dplyr::select(-a) %>%
dplyr::rename(dependson = b) #%>%
j %>% filter(!is.na(dependson)) %>% select(dependson)# A tibble: 37 × 1
dependson
<chr>
1 "the design of course"
2 "the topic"
3 "the study. if you have interstimulus interval of 1 second and you expect to…
4 "the experiment and research question"
5 "the rest period between the measured evoked responses. e.g. it can be very …
6 "the analysis"
7 "paradigm, 100-300 ms range preferable"
8 "the task design"
9 "the type of response (eg for mrcps response is seen before actual movement …
10 "the task design, paradigm and signal of interest."
# ℹ 27 more rows# A tibble: 21 × 1
q
<chr>
1 minimum 200ms for erps and theta or beta power
2 should match the duration of baseline-correction
3 as a rule of thumb, i would plot at least 1/3 of the duration (post-stimulus…
4 half of the illustrated task interval
5 in general i would always try to plot the full baseline period used for base…
6 the same duration as the one used for baseline correction
7 at least 300, preferably more
8 put down 100, but that's just what i typically use, might be diff for differ…
9 at least the baseline window used for the baseline correction?
10 in this case it has sense as the -100 : 0 ms is not flat
# ℹ 11 more rowsbl <- table(abs(just[2] %>% na.omit() %>% rbind(data[77] %>% dplyr::rename(num = !!names(.)[1]) , .))) %>% data.frame() %>% dplyr::rename(baseline = !!names(.)[1]) %>%
mutate(percent_score = round(Freq / sum(Freq), 2) *100)
periods_plot <- bl %>%
ggplot(data = ., aes(x = baseline, y = percent_score)) +
geom_bar(stat="identity", fill ="lightblue1", colour ="dodgerblue3") +
labs(x = "Recomended baseline duration in msec", y = "") +
scale_y_continuous(breaks=seq(0, 60, 5)) + theme_classic() +
theme(legend.position="none", plot.title = element_text(hjust = 0.5), text = element_text(family = "Lato")) +
geom_text(aes(label = paste0(percent_score, "%"), group = baseline), position = position_dodge(width = .9), vjust = -0.3) +
theme(legend.position="none", plot.caption = element_text(hjust=0), axis.text.y = element_blank(), axis.text = element_text(size = 10))
periods_plot +
labs(caption = sprintf("N of respondents - %d", sum(bl$Freq)))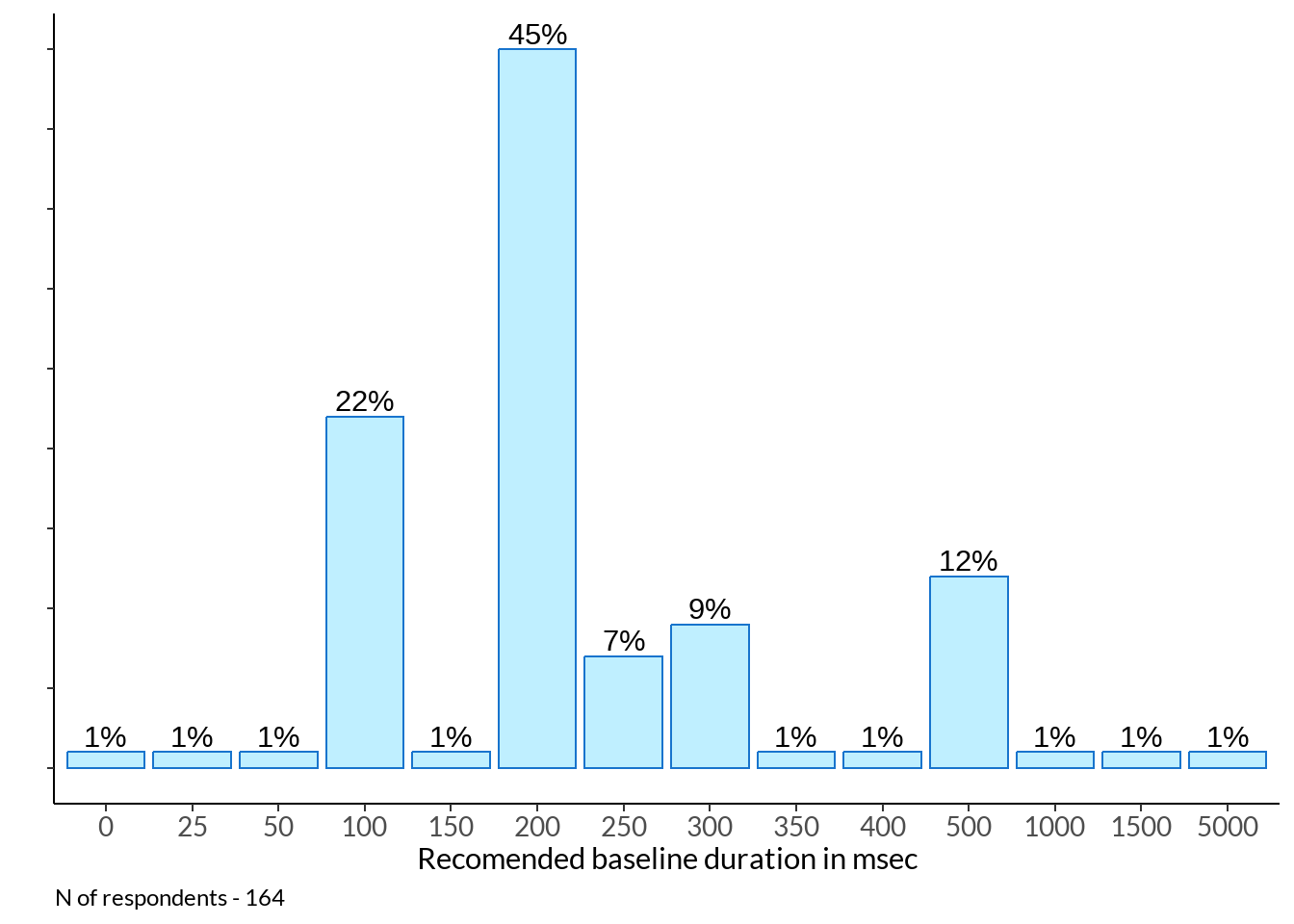
bl2 <- table(abs(just[2] %>% na.omit() %>% rbind(data[77] %>% dplyr::rename(num = !!names(.)[1]) , .))) %>% data.frame() %>% dplyr::rename(baseline = !!names(.)[1]) %>%
mutate(baseline = ifelse(Freq > 2, paste0("-", as.character(baseline)), "Other")) %>%
dplyr::group_by(baseline) %>% dplyr::summarise(Freq = sum(Freq)) %>%
mutate(percent_score = round(Freq / sum(Freq), 2) *100) %>% arrange(desc(baseline)) %>%
mutate(baseline = factor(baseline,
levels = c("-500", "-300", "-250", "-200", "-100", "Other")))
italised1 <- c("-500", "-300", "-250", "-200", "-100", expression(italic("Other")))
periods_plot2 <- bl2 %>%
ggplot(data = ., aes(x = baseline, y = percent_score)) +
geom_bar(stat="identity", fill ="#6BAED6", width = 0.6) +
labs(x = "Recomended baseline duration\n(msec)", y = "") +
scale_y_continuous(breaks=seq(0, 60, 5)) + theme_classic() +
theme(legend.position="none", plot.title = element_text(hjust = 0.5), text = element_text(family = "Lato")) +
geom_text(aes(label = paste0(percent_score, "%"), group = baseline), position = position_dodge(width = .9), vjust = -0.3) +
theme(legend.position="none", plot.caption = element_text(hjust=0), axis.text.y = element_blank(), axis.text = element_text(size = 10)) +
scale_x_discrete(labels = italised1)
periods_plot2 +
labs(caption = sprintf("N of respondents - %d", sum(bl$Freq)))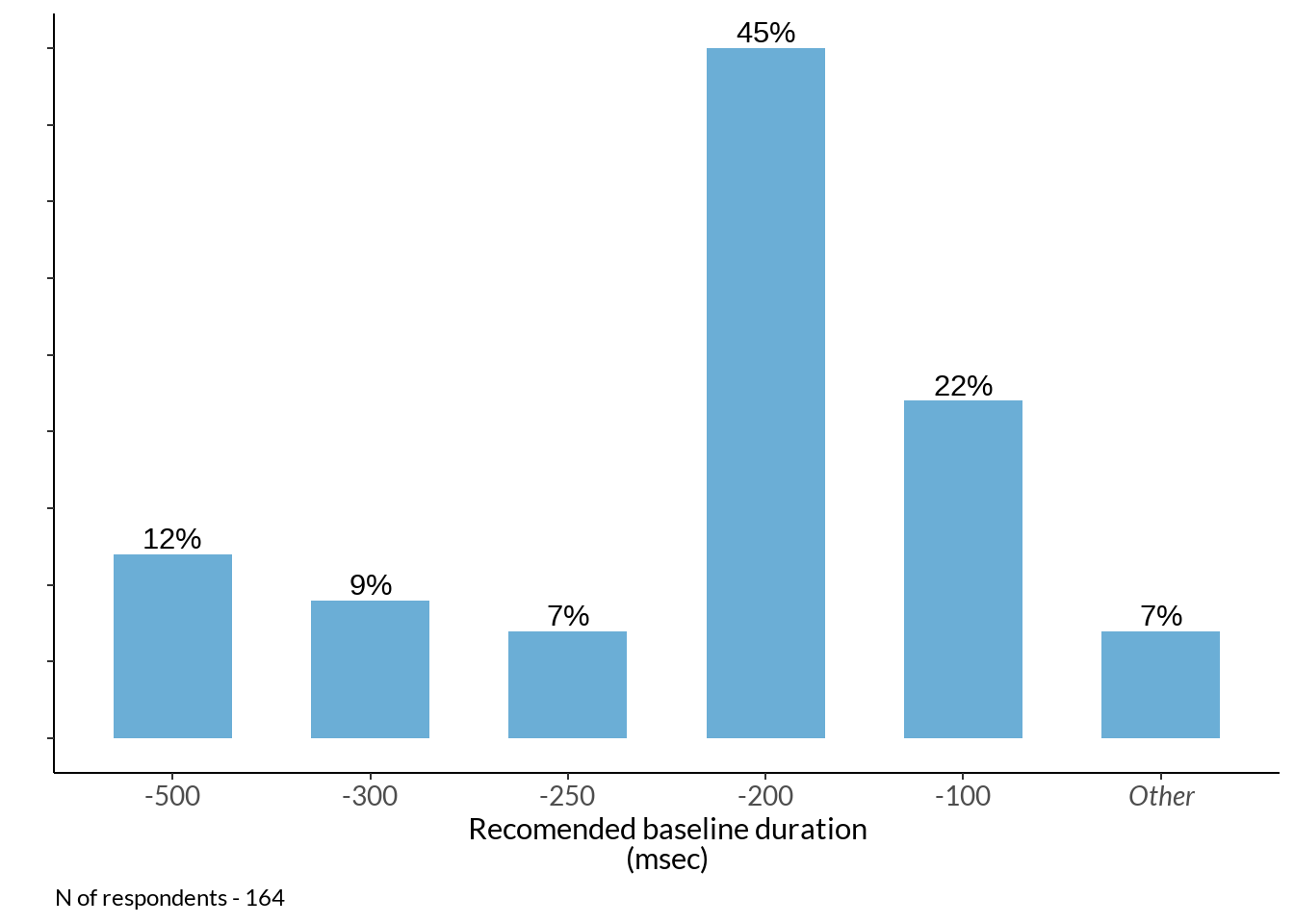
tti_data <- data[95] %>% rename_at(vars(colnames(.)), ~ c("toposeries_int")) %>% filter(!is.na(.), toposeries_int != "Other") %>%
group_by(toposeries_int) %>% dplyr::summarise(n = n()) %>% mutate(toposeries_int = case_when(
toposeries_int == "The instantaneous time slices (single time points)" ~ "Time point",
toposeries_int == "The mean value of time bin, centered around the labelled time (average over multiple time points)"~ "Average over\ntime points"
)) %>% mutate(p = round(n / sum(n) * 100))
tti <- tti_data %>% ggplot(., aes(y = toposeries_int, x = p)) +
geom_bar(stat = "identity", width=0.5, position = "dodge", fill ="#6BAED6") + theme_classic() +
theme(
text = element_text(family = "Lato"),
legend.position="none",
strip.background = element_blank(),
axis.ticks = element_line(color = "grey40", size = .5),
axis.text.x = element_blank(),
axis.text.y = element_text(color = "grey10", size = 16),
axis.title.x=element_blank(),
plot.title = element_text(color = "grey10", size = 18, face = "bold", margin = margin(t = 15)),
plot.title.position = "plot",
plot.caption = element_text(hjust=0),
) +
geom_text(aes(label = paste0(p, "%"), group = toposeries_int), hjust = -0.2, size = 5, family = "Lato") +
labs(title = "How do you interpret time in topoplot timeseries?", subtitle = "", y = "")+
scale_fill_manual(values=colours) +
xlim(0, 100)
tti +
labs(caption = sprintf("N of respondents - %d", sum(tti_data$n))) # change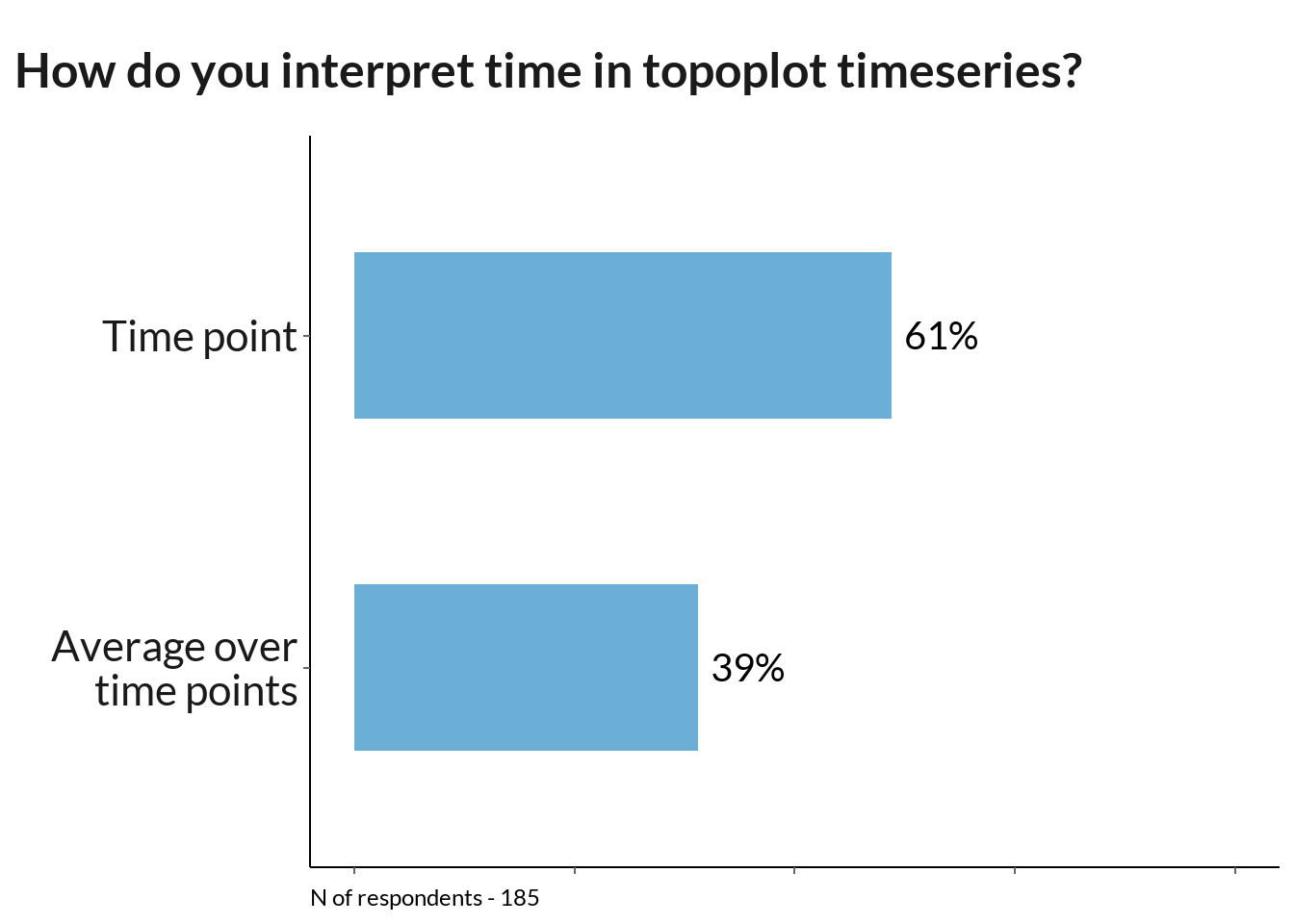
position Scores sum
1 No 63 157
2 Yes 94 157table(data[74]) %>% data.frame() %>% rename_at(vars(colnames(.)), ~ c("position", "Scores")) %>% mutate(percent_score = round(Scores / sum(Scores) * 100)) %>%
ggplot(., aes(x = position, y = percent_score, fill = as.factor(percent_score))) +
geom_bar(position = "dodge", stat = "identity", width=0.5) + theme_classic() +
theme(axis.title.x=element_blank(),
legend.position="none",
axis.text = element_text(size = 12),
plot.title = element_text(hjust = 0.5)) +
geom_text(aes(label = paste0(percent_score, "%"), group = position), position = position_stack(vjust = 0.5), size = 6) +
ggtitle("In your paper, did you published ERP plot with error bars?")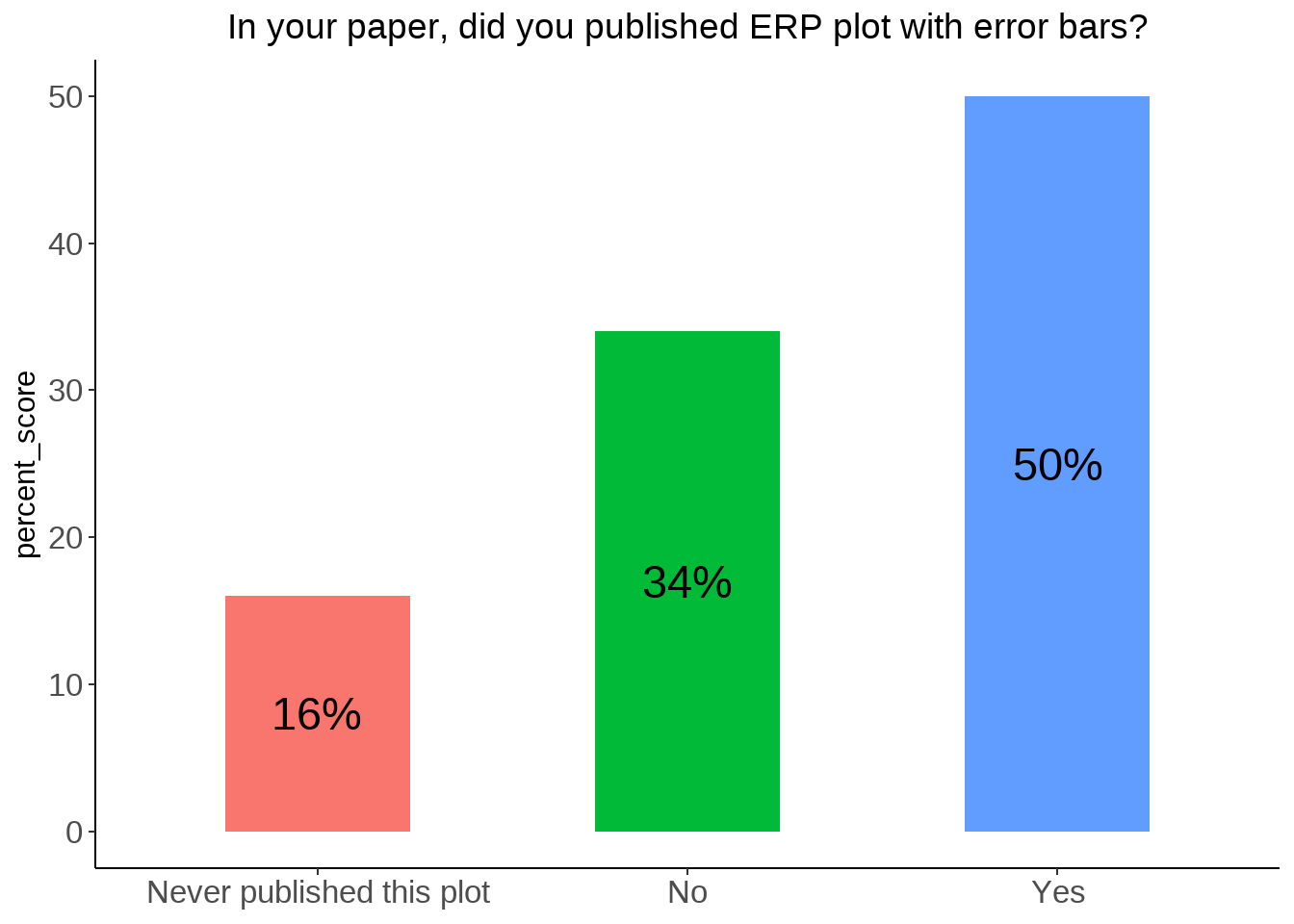
eb <- table(data[74]) %>% data.frame() %>% rename_at(vars(colnames(.)), ~ c("Position", "Scores")) %>%
filter(Position != "Never published this plot") %>%
mutate(percent_score = round(Scores / sum(Scores) * 100))
eb_fig <- eb %>%
ggplot(., aes(y = Position, x = percent_score)) +
geom_bar(stat = "identity", width=0.5, position = "dodge", fill ="#6BAED6") + theme_classic() +
theme(axis.text.x = element_blank(),
axis.title.y = element_blank(),
legend.position="none",
text = element_text(family = "Lato"),
axis.text = element_text(size = 12),
plot.title = element_text(hjust = 0.5)) +
geom_text(aes(label = paste0(percent_score, "%"), group = Position), #position = position_stack(vjust = 0.5),
hjust = -0.2) +
ggtitle("In your paper, have you published\nan ERP plot with error bars?")+
scale_fill_manual(values=colours) +
theme(legend.position="none", plot.caption = element_text(hjust=0), axis.text = element_text(size = 10)) +
xlim(0, 100) + labs(y = "")
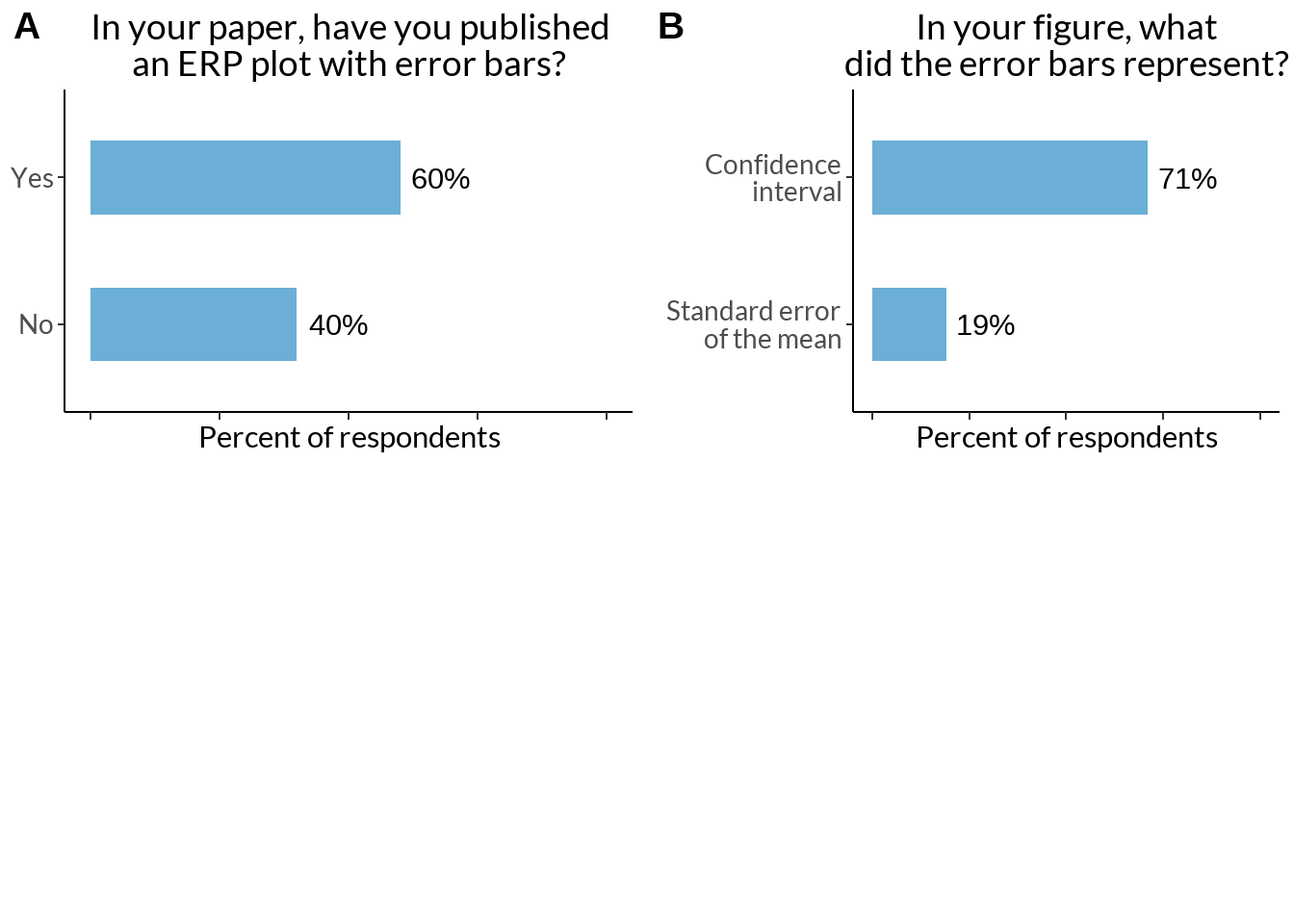
eb_fig +
labs(caption = sprintf("N of respondents - %d", sum(eb$Scores)))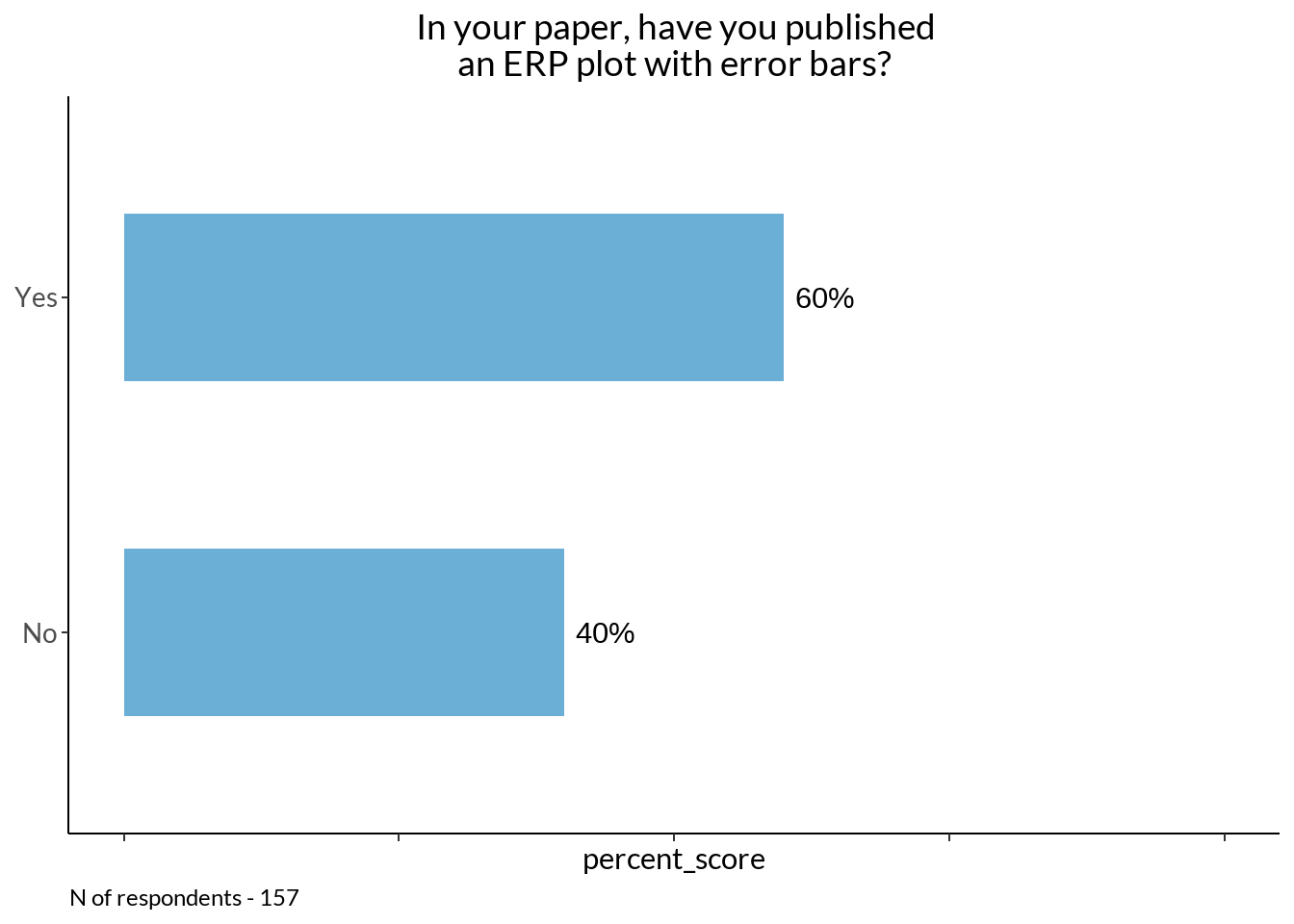
ebd <- data[75] %>% filter(!is.na(.)) %>% table() %>% data.frame() %>% rename_at(vars(colnames(.)), ~ c("position", "scores")) %>% mutate(percent_score = round(scores / sum(scores) * 100)) %>% filter(position != "Other")
ebd_fig <- ebd %>%
ggplot(., aes(y = position, x = percent_score)) +
geom_bar(position = "dodge", stat = "identity", width=0.5, fill ="#6BAED6") +
theme_classic() +
theme(axis.text.x = element_blank(),
text = element_text(family = "Lato"),
legend.position="none",
axis.title.y = element_blank(),
plot.title = element_text(hjust = 0.5)) +
geom_text(aes(label = paste0(percent_score, "%"), group = position), hjust = -0.2) +
ggtitle("In your figure, what\ndid the error bars represent?")+
scale_fill_manual(values=colours) + scale_y_discrete(labels = c("Standard error\nof the mean", "Confidence\ninterval")) +
theme(plot.caption = element_text(hjust=0), axis.text = element_text(size = 10)) + xlim(0, 100)
ebd_fig +
labs(caption = sprintf("N of respondents - %d", sum(ebd$scores))) 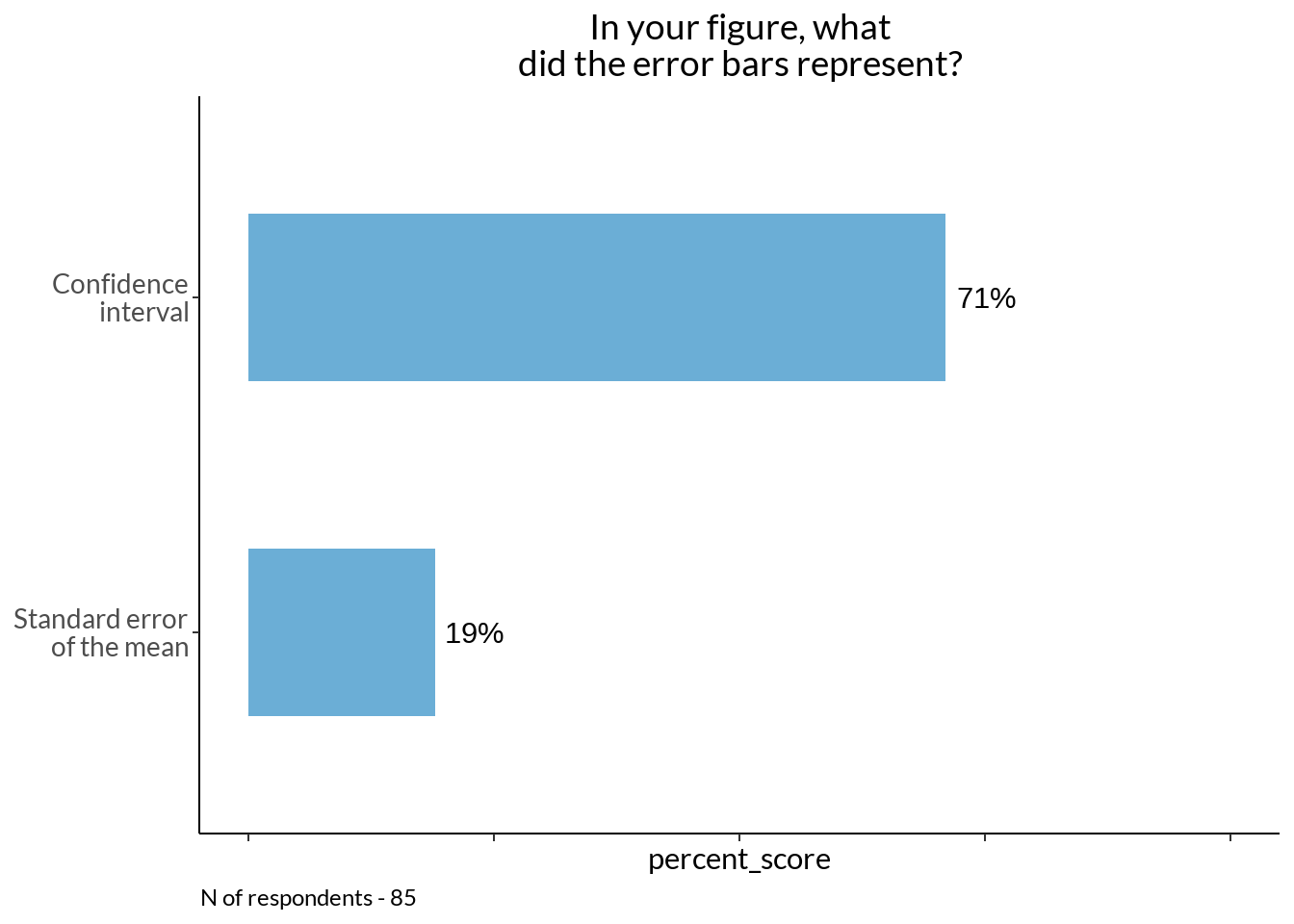
G03Q15[other]. What did the error bars depict in your figure? [Other]
-
1
68% CI, which is close to SEM under normality
1
95% ci over channel means
1
i'm not sure but i think it was sem
1
I don't remember
1
median absolute deviaton or quantiles
1
NA
1
Sd
1
SEM, corrected for within-participant design
1 cb <- table(data[117]) %>% data.frame() %>% rename_at(vars(colnames(.)), ~ c("position", "scores"))
cb %>%
mutate(percent_score = round(scores / sum(scores) * 100)) %>%
ggplot(., aes(y = position, x = percent_score)) + #, fill = as.factor(percent_score))) +
geom_bar(position = "dodge", stat = "identity", width=0.5, fill ="#6BAED6") +
theme_classic() +
theme(
text = element_text(family = "Lato"),
legend.position="none",
strip.background = element_blank(),
axis.title.y = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_text(color = "grey10", size = 16),
axis.title.x = element_text(color = "grey10", size = 16),
plot.title = element_text(color = "grey10", size = 18, face = "bold", margin = margin(t = 15)),
plot.title.position = "plot",
plot.caption = element_text(hjust=0),
) + xlab("Percent of respondents") +
geom_text(aes(label = paste0(percent_score, "%"), group = position), hjust = -0.2, size = 7) +
ggtitle("Are you aware about\nperceptual controvercies of colormaps?")+
scale_fill_manual(values=colours) +
theme(legend.position="none", plot.caption = element_text(hjust=0)) + xlim(0, 65) #+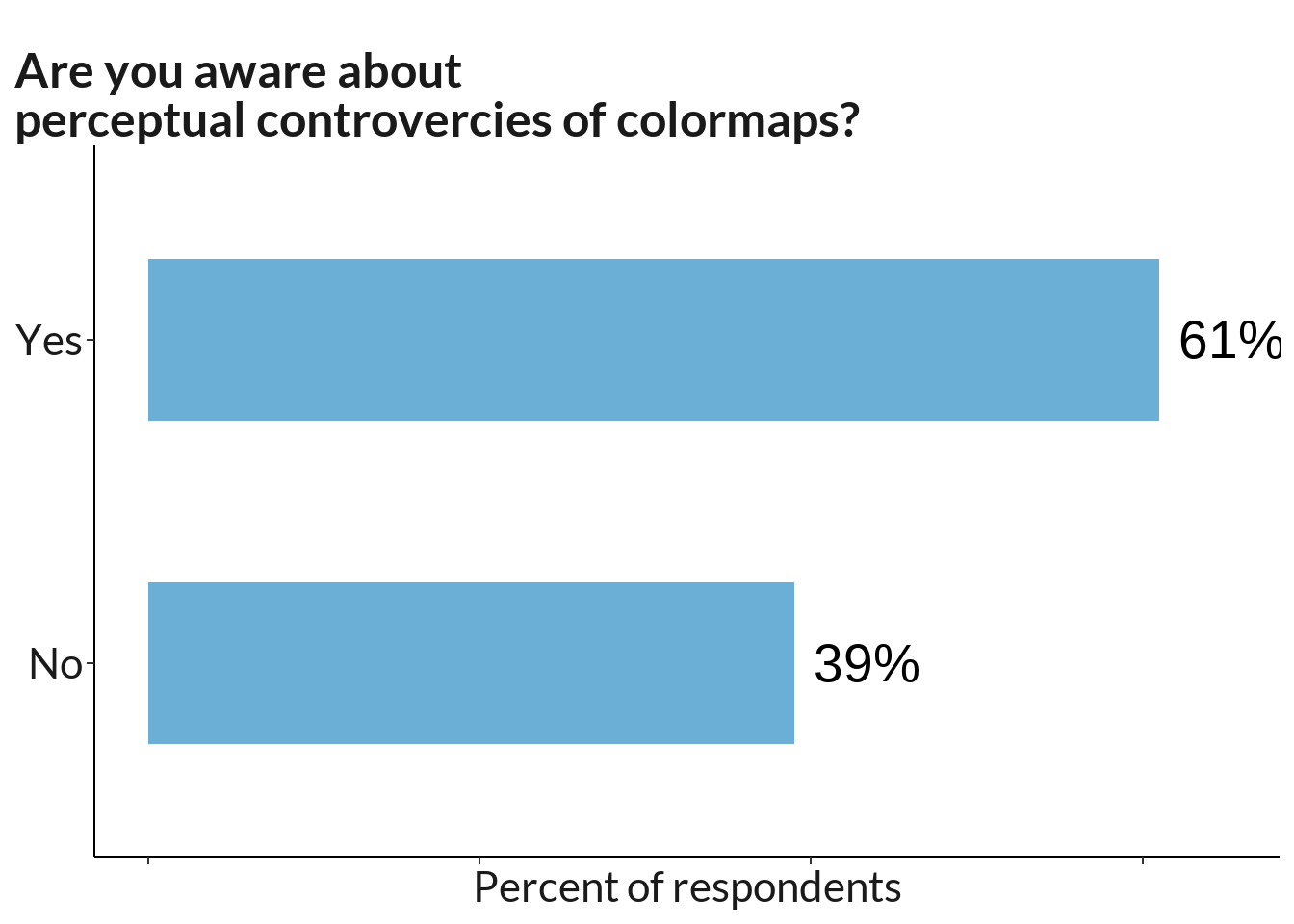
$caption
[1] "N of respondents - 213"
attr(,"class")
[1] "labels"rbind(table(data[117]) %>% data.frame() %>%
rename_at(vars(colnames(.)), ~ c("answer", "scores")) %>%
mutate(questions = "Awareness about\ncontrovercies of\ncolorbars")%>%
mutate(percent_score = round(scores / sum(scores) * 100)),
table(data[118]) %>% data.frame() %>%
rename_at(vars(colnames(.)), ~ c("answer", "scores")) %>%
mutate(questions = "Awareness about\n2D colorbars") %>%
mutate(percent_score = round(scores / sum(scores) * 100))
) %>%
ggplot(., aes(x = percent_score, y = questions, fill = answer)) +
geom_col(stat = "identity", width = 0.5) +
geom_text(aes(label = paste0(percent_score, "%", "\n(", answer, ")")),
position = position_stack(vjust = 0.5), size = 4) +
theme_classic()+
theme(plot.title = element_text(hjust = 0.5),
axis.text = element_text(size = 12),
axis.title = element_text(size = 14, face = "bold"),
legend.position = "none",
legend.title = element_blank(),
axis.title.y=element_blank()
) +
scale_color_manual(values = c("#FF6666", "#66CC66")) +
scale_fill_manual(values = c("#FF6666", "#66CC66")) +
labs(x="Percent score") +
labs(caption = sprintf("N of respondents - %d", sum(cb$scores))) +
theme(legend.position="none", plot.caption.position = "plot", plot.caption = element_text(hjust=0), axis.text = element_text(size = 10)) 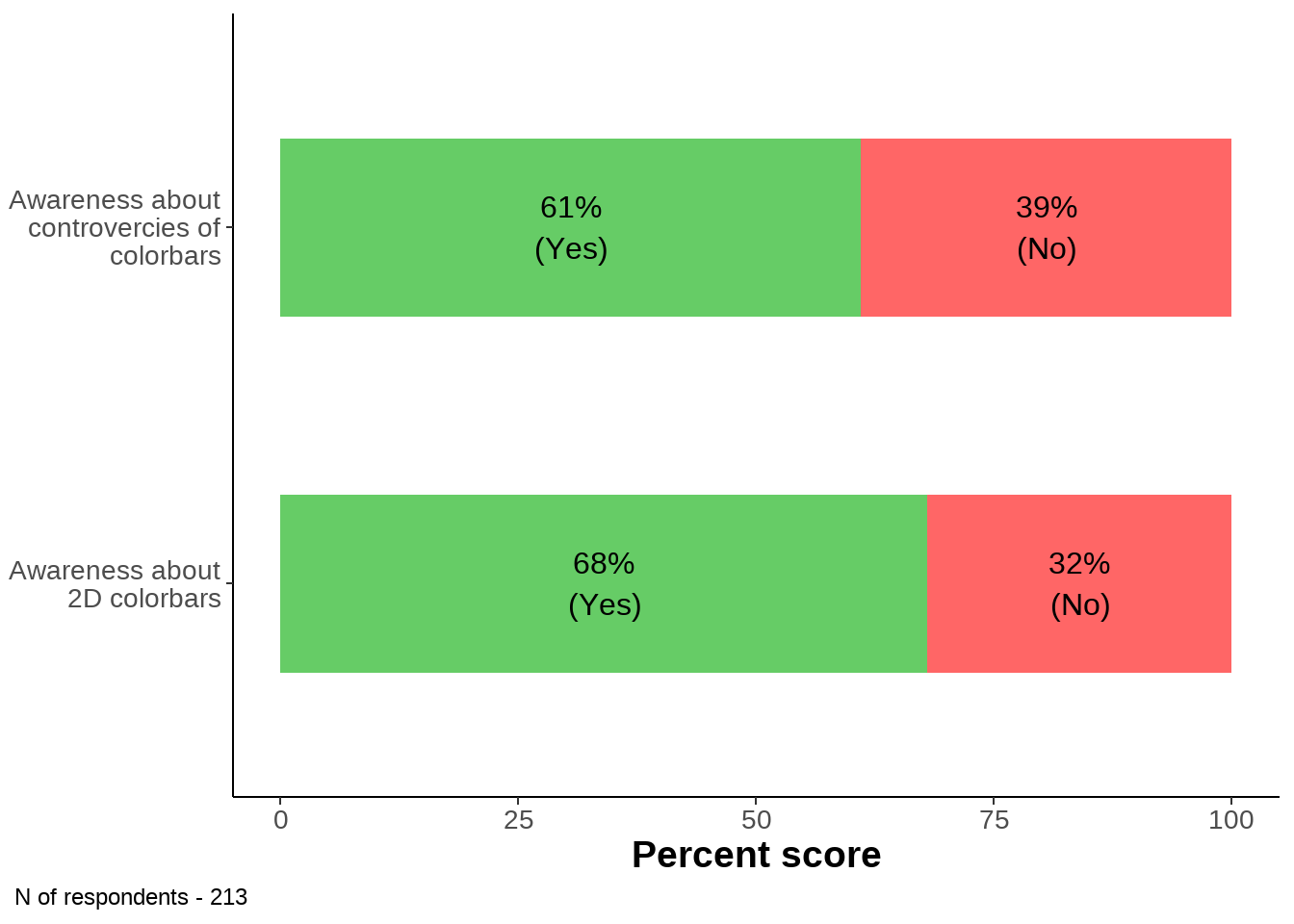
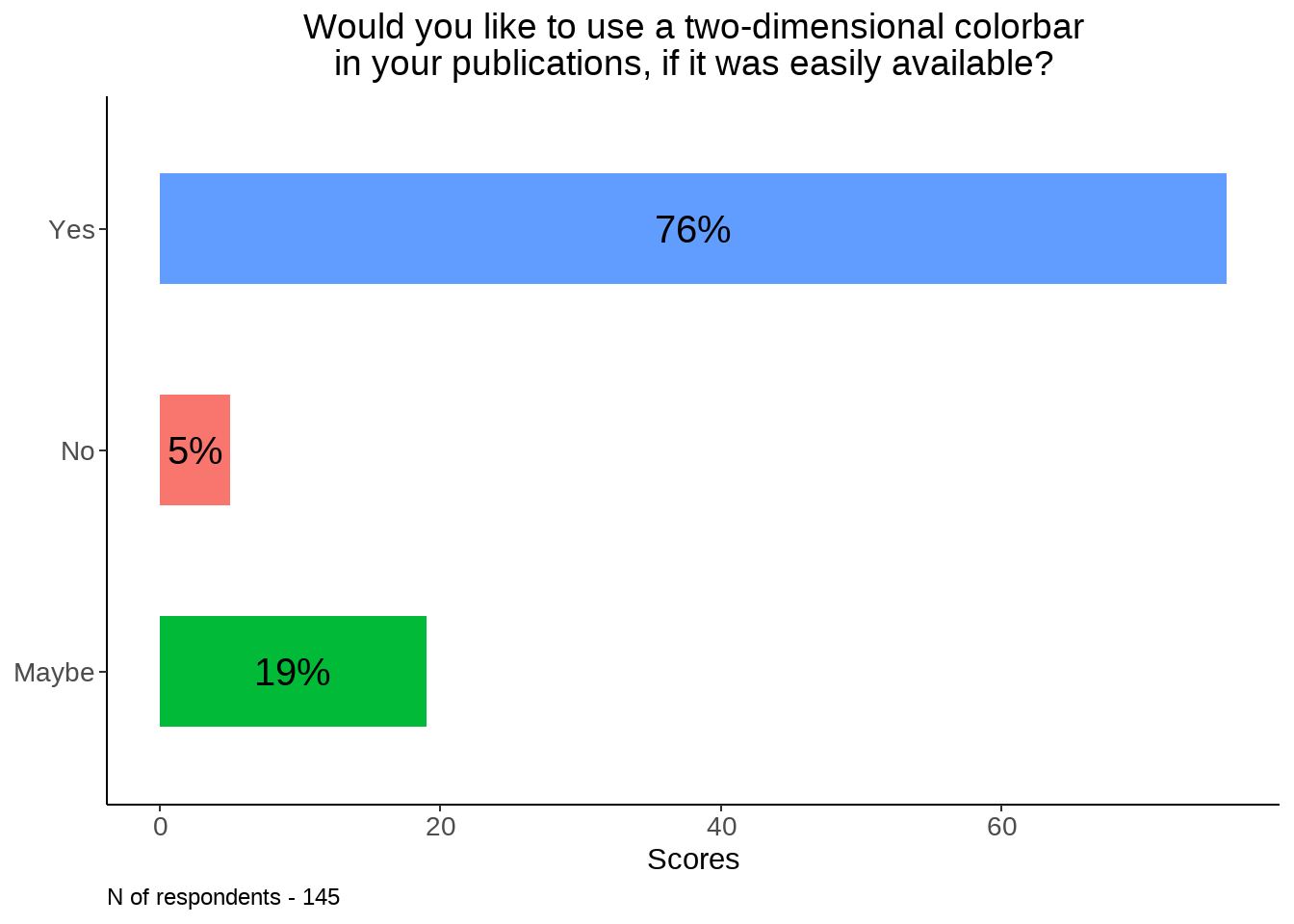
tdc <- table(data[119]) %>% data.frame() %>% rename_at(vars(colnames(.)), ~ c("position", "scores")) %>%
mutate(percent_score = round(scores / sum(scores) * 100))
tdc %>%
ggplot(., aes(y = position, x = percent_score, fill = as.factor(percent_score))) +
geom_bar(stat = "identity", width=0.5) + theme_classic() +
theme(axis.title.y=element_blank(),
legend.position="none",
axis.text = element_text(size = 12),
plot.title = element_text(hjust = 0.5)) + labs(x = "Scores") +
geom_text(aes(label = paste0(percent_score, "%") ,
group = position), position = position_stack(vjust = 0.5), size = 5) +
ggtitle("Would you like to use a two-dimensional colorbar\nin your publications, if it was easily available?") +
labs(caption = sprintf("N of respondents - %d", sum(tdc$scores))) +
theme(legend.position="none", plot.caption = element_text(hjust=0), axis.text = element_text(size = 10))